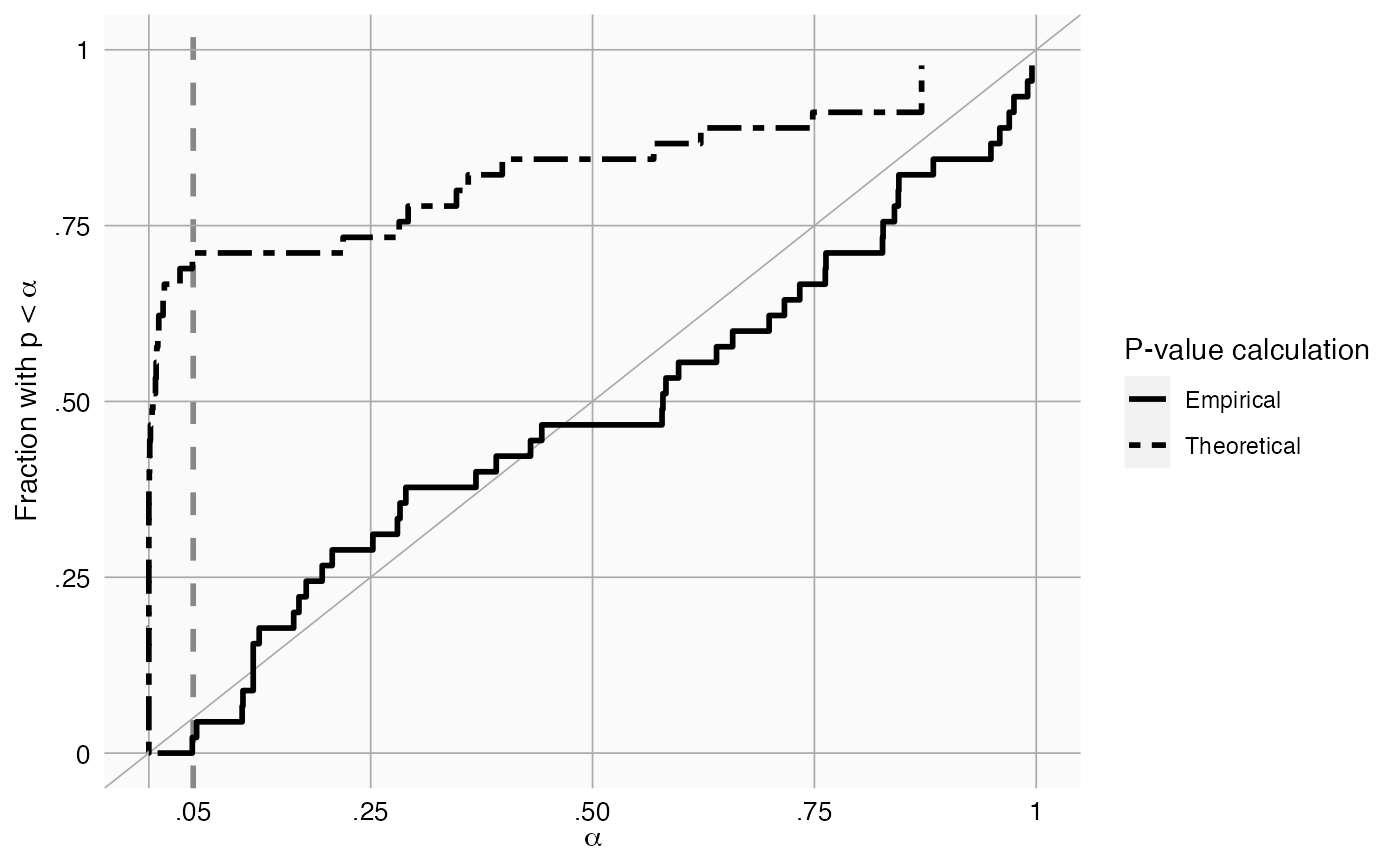
plotCalibration creates a plot showing the calibration of our calibration procedure
plotCalibration(
logRr,
seLogRr,
useMcmc = FALSE,
legendPosition = "right",
title,
fileName = NULL
)Arguments
- logRr
A numeric vector of effect estimates on the log scale
- seLogRr
The standard error of the log of the effect estimates. Hint: often the standard error = (log(<lower bound 95 percent confidence interval>) - log(<effect estimate>))/qnorm(0.025)
- useMcmc
Use MCMC to estimate the calibrated P-value?
- legendPosition
Where should the legend be positioned? ("none", "left", "right", "bottom", "top")
- title
Optional: the main title for the plot
- fileName
Name of the file where the plot should be saved, for example 'plot.png'. See the function
ggsavein the ggplot2 package for supported file formats.
Value
A Ggplot object. Use the ggsave function to save to file.
Details
Creates a calibration plot showing the number of effects with p < alpha for every level of alpha. The empirical calibration is performed using a leave-one-out design: The p-value of an effect is computed by fitting a null using all other negative controls. Ideally, the calibration line should approximate the diagonal. The plot shows both theoretical (traditional) and empirically calibrated p-values.
Examples
data(sccs)
negatives <- sccs[sccs$groundTruth == 0, ]
plotCalibration(negatives$logRr, negatives$seLogRr)