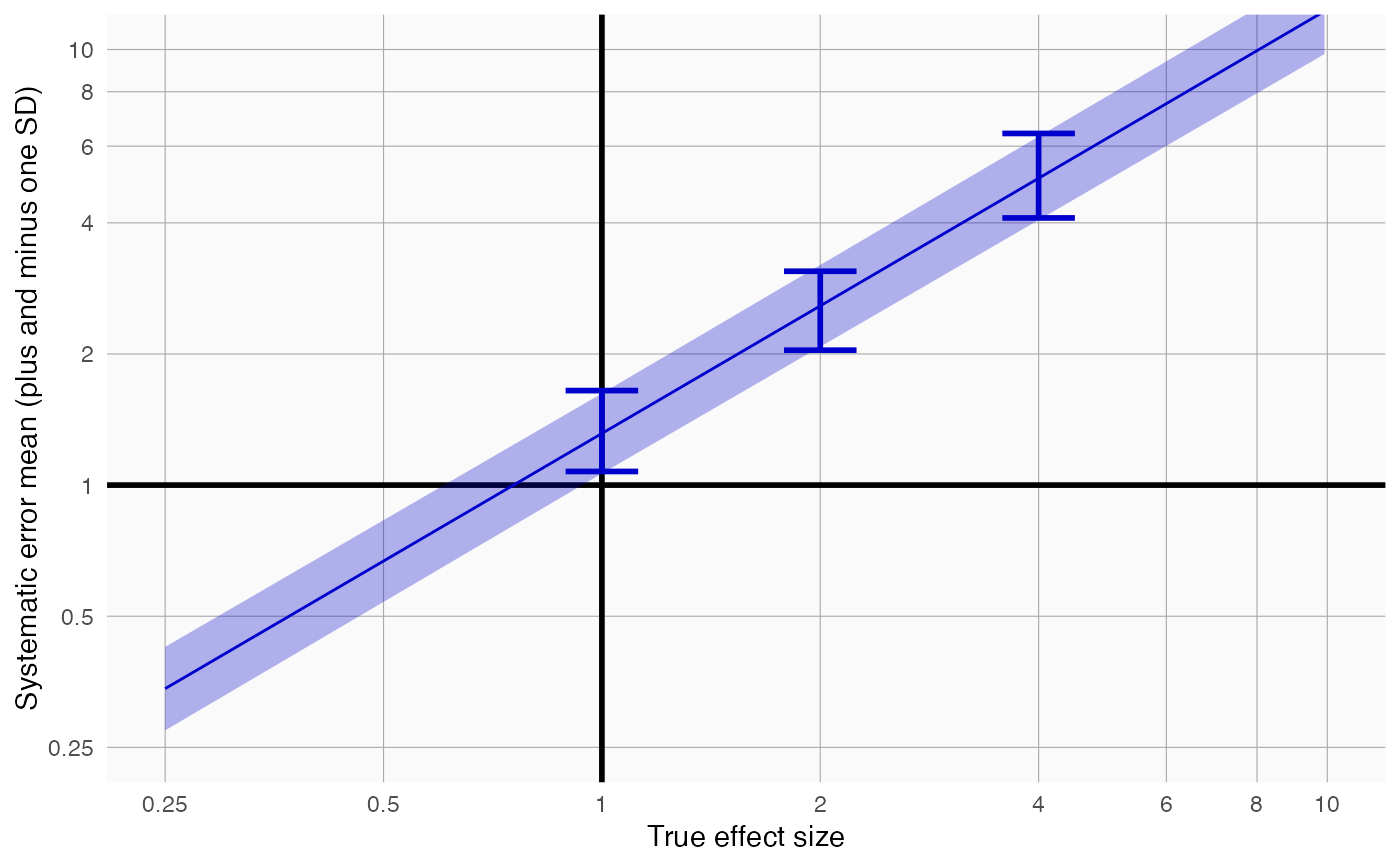
plotErrorModel creates a plot showing the systematic error model.
plotErrorModel(
logRr,
seLogRr,
trueLogRr,
title,
legacy = FALSE,
fileName = NULL
)Arguments
- logRr
A numeric vector of effect estimates on the log scale.
- seLogRr
The standard error of the log of the effect estimates. Hint: often the standard error = (log(<lower bound 95 percent confidence interval>) - log(<effect estimate>))/qnorm(0.025).
- trueLogRr
The true log relative risk.
- title
Optional: the main title for the plot
- legacy
If true, a legacy error model will be fitted, meaning standard deviation is linear on the log scale. If false, standard deviation is assumed to be simply linear.
- fileName
Name of the file where the plot should be saved, for example 'plot.png'. See the function
ggsavein the ggplot2 package for supported file formats.
Value
A Ggplot object. Use the ggsave function to save to file.
Details
Creates a plot with the true effect size on the x-axis, and the mean plus and minus the standard deviation shown on the y-axis. Also shown are simple error models fitted at each true relative risk in the input.
Examples
data <- simulateControls(n = 50 * 3, mean = 0.25, sd = 0.25, trueLogRr = log(c(1, 2, 4)))
plotErrorModel(data$logRr, data$seLogRr, data$trueLogRr)