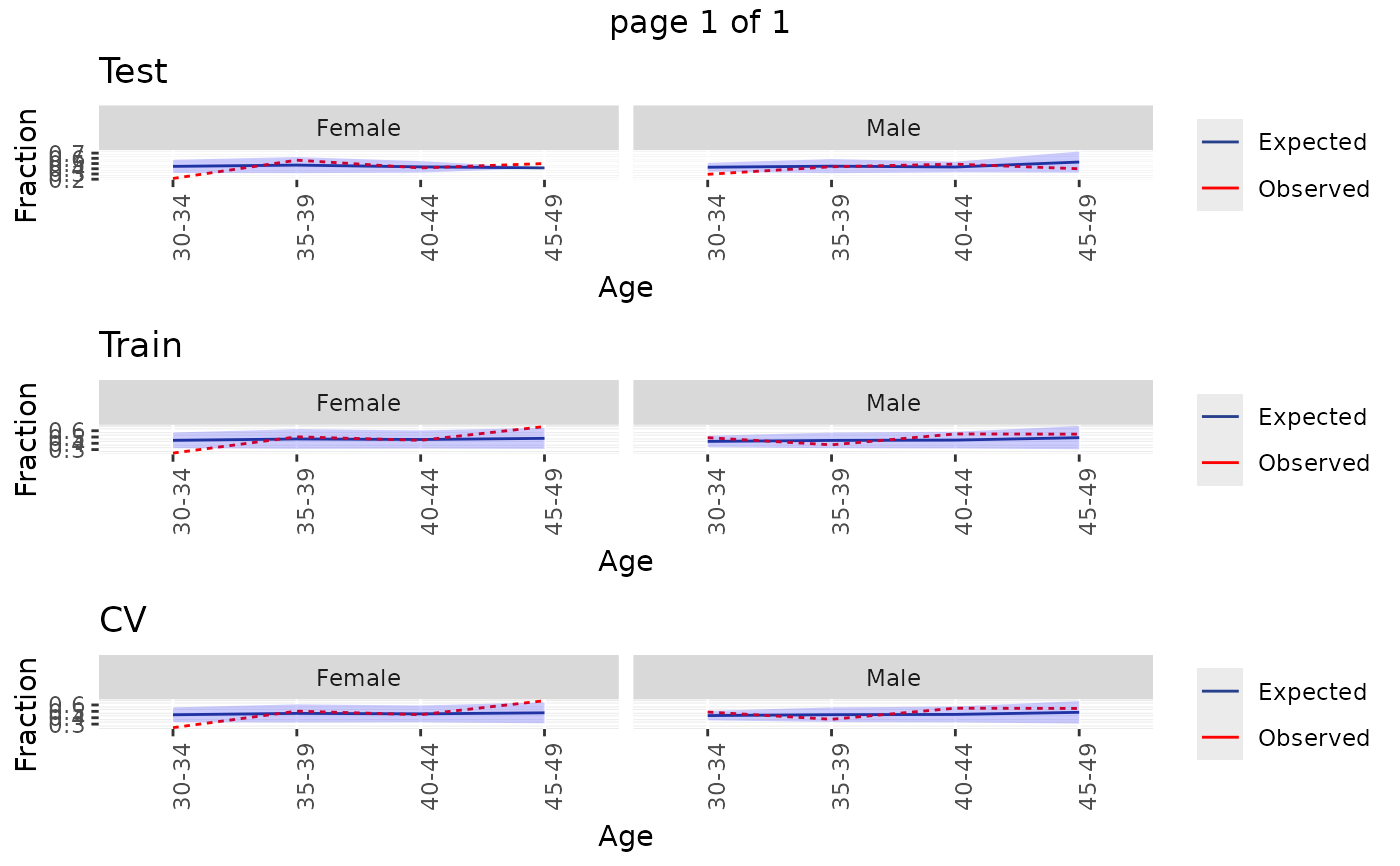
Plot the Observed vs. expected incidence, by age and gender
Source:R/Plotting.R
plotDemographicSummary.RdPlot the Observed vs. expected incidence, by age and gender
Usage
plotDemographicSummary(
plpResult,
typeColumn = "evaluation",
saveLocation = NULL,
fileName = "roc.png"
)Arguments
- plpResult
A plp result object as generated using the
runPlpfunction.- typeColumn
The name of the column specifying the evaluation type
- saveLocation
Directory to save plot (if NULL plot is not saved)
- fileName
Name of the file to save to plot, for example 'plot.png'. See the function
ggsavein the ggplot2 package for supported file formats.
Value
A ggplot object. Use the ggsave function to save to file in a different
format.
Examples
# \donttest{
data("simulationProfile")
plpData <- simulatePlpData(simulationProfile, n = 1000, seed = 42)
#> Generating covariates
#> Generating cohorts
#> Generating outcomes
saveLoc <- file.path(tempdir(), "plotDemographicSummary")
plpResult <- runPlp(plpData, outcomeId = 3, saveDirectory = saveLoc)
#> Use timeStamp: TRUE
#> Creating save directory at: /tmp/RtmpBBYORx/plotDemographicSummary/2025-07-25-3
#> Currently in a tryCatch or withCallingHandlers block, so unable to add global calling handlers. ParallelLogger will not capture R messages, errors, and warnings, only explicit calls to ParallelLogger. (This message will not be shown again this R session)
#> Patient-Level Prediction Package version 6.4.1.9999
#> Study started at: 2025-07-25 10:04:36.665044
#> AnalysisID: 2025-07-25-3
#> AnalysisName: Study details
#> TargetID: 1
#> OutcomeID: 3
#> Cohort size: 1000
#> Covariates: 98
#> Creating population
#> Outcome is 0 or 1
#> Population created with: 954 observations, 954 unique subjects and 434 outcomes
#> Population created in 0.0491 secs
#> seed: 123
#> Creating a 25% test and 75% train (into 3 folds) random stratified split by class
#> Data split into 238 test cases and 716 train cases (239, 239, 238)
#> Data split in 0.796 secs
#> Train Set:
#> Fold 1 239 patients with 109 outcomes - Fold 2 239 patients with 109 outcomes - Fold 3 238 patients with 108 outcomes
#> 66 covariates in train data
#> Test Set:
#> 238 patients with 108 outcomes
#> Removing 2 redundant covariates
#> Removing 0 infrequent covariates
#> Normalizing covariates
#> Tidying covariates took 1.16 secs
#> Train Set:
#> Fold 1 239 patients with 109 outcomes - Fold 2 239 patients with 109 outcomes - Fold 3 238 patients with 108 outcomes
#> 64 covariates in train data
#> Test Set:
#> 238 patients with 108 outcomes
#>
#> Running Cyclops
#> Done.
#> GLM fit status: OK
#> Creating variable importance data frame
#> Prediction took 0.155 secs
#> Time to fit model: 0.311 secs
#> Removing infrequent and redundant covariates and normalizing
#> Removing infrequent and redundant covariates covariates and normalizing took 0.36 secs
#> Prediction took 0.156 secs
#> Prediction done in: 0.876 secs
#> Calculating Performance for Test
#> =============
#> AUC 61.40
#> 95% lower AUC: 54.59
#> 95% upper AUC: 68.21
#> AUPRC: 57.58
#> Brier: 0.23
#> Eavg: 0.06
#> Calibration in large- Mean predicted risk 0.4497 : observed risk 0.4538
#> Calibration in large- Intercept 0.3069
#> Weak calibration intercept: 0.3069 - gradient:2.3474
#> Hosmer-Lemeshow calibration gradient: 2.41 intercept: -0.69
#> Average Precision: 0.58
#> Calculating Performance for Train
#> =============
#> AUC 59.21
#> 95% lower AUC: 55.21
#> 95% upper AUC: 63.20
#> AUPRC: 55.33
#> Brier: 0.24
#> Eavg: 0.03
#> Calibration in large- Mean predicted risk 0.4553 : observed risk 0.4553
#> Calibration in large- Intercept 0.0515
#> Weak calibration intercept: 0.0515 - gradient:1.2817
#> Hosmer-Lemeshow calibration gradient: 1.31 intercept: -0.14
#> Average Precision: 0.55
#> Calculating Performance for CV
#> =============
#> AUC 56.25
#> 95% lower AUC: 52.05
#> 95% upper AUC: 60.46
#> AUPRC: 52.52
#> Brier: 0.24
#> Eavg: 0.02
#> Calibration in large- Mean predicted risk 0.4558 : observed risk 0.4553
#> Calibration in large- Intercept 0.0403
#> Weak calibration intercept: 0.0403 - gradient:1.2344
#> Hosmer-Lemeshow calibration gradient: 1.19 intercept: -0.09
#> Average Precision: 0.53
#> Time to calculate evaluation metrics: 0.216 secs
#> Calculating covariate summary @ 2025-07-25 10:04:40.367607
#> This can take a while...
#> Creating binary labels
#> Joining with strata
#> calculating subset of strata 1
#> calculating subset of strata 2
#> calculating subset of strata 3
#> calculating subset of strata 4
#> Restricting to subgroup
#> Calculating summary for subgroup TrainWithOutcome
#> Restricting to subgroup
#> Calculating summary for subgroup TrainWithNoOutcome
#> Restricting to subgroup
#> Calculating summary for subgroup TestWithOutcome
#> Restricting to subgroup
#> Calculating summary for subgroup TestWithNoOutcome
#> Aggregating with labels and strata
#> Finished covariate summary @ 2025-07-25 10:04:43.822783
#> Time to calculate covariate summary: 3.46 secs
#> Run finished successfully.
#> Saving PlpResult
#> Creating directory to save model
#> plpResult saved to ..\/tmp/RtmpBBYORx/plotDemographicSummary/2025-07-25-3\plpResult
#> runPlp time taken: 7.25 secs
plotDemographicSummary(plpResult)
 # clean up
unlink(saveLoc, recursive = TRUE)
# }
# clean up
unlink(saveLoc, recursive = TRUE)
# }