Evaluation of the exact matching pipeline¶
Here we evaluate the performance on the task of finding exact matches of source terms to standard concepts in the OHDSI Vocabulary. For this we use a gold standard dataset of source terms with their correct mappings to standard concepts, with some source terms not having any correct mapping.
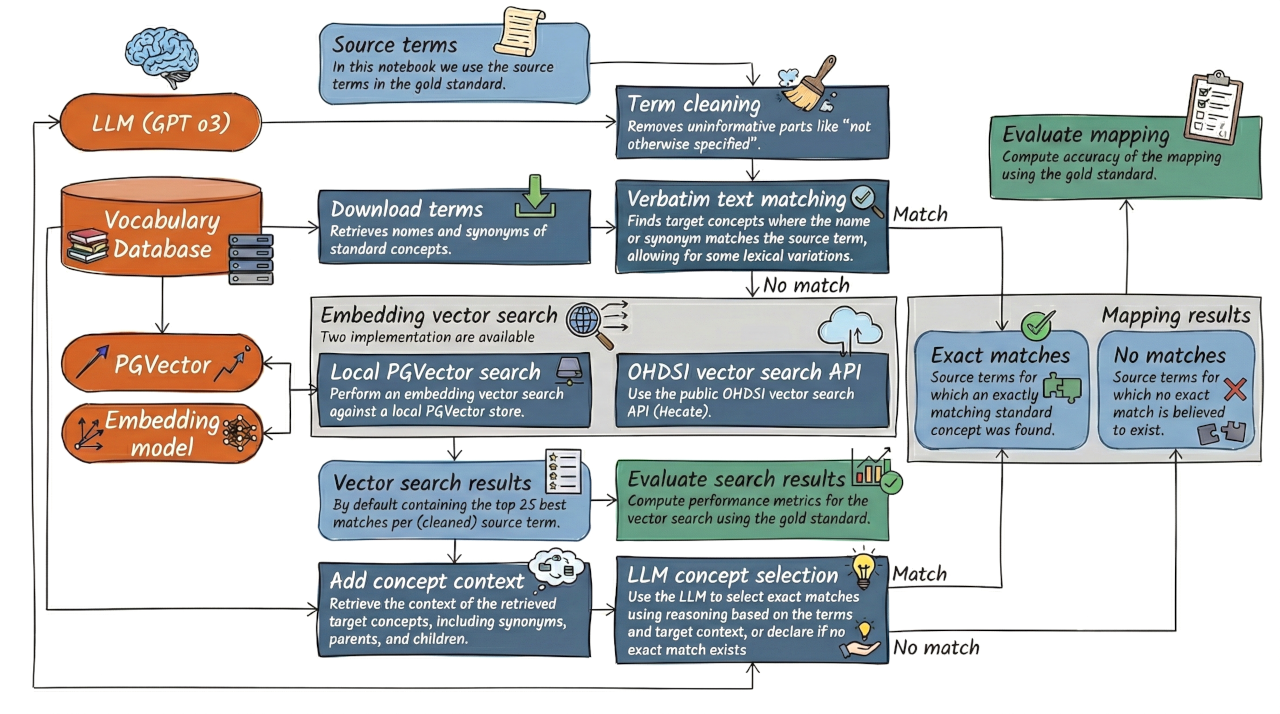
Setup¶
Before running this notebook, make sure the environment is set up as described in the README.
This includes credentials for a database with the OHDSI Vocabulary loaded, and credentials for the LLM API.
The prompts, as specified in config.yaml, are tailored for GPT o3, so we recommend using that model for this evaluation.
This notebook currently assumes the use of a local PGVector instance for vector search, instantiated with the code at https://github.com/schuemie/OhdsiVocabVectorStore, but an option to use the public OHDSI Hecate vector search API is also available.
All results will be stored in the data/notebook_results folder.
Most code blocks will load results from file if they already exist, to save time and costs.
Delete those files to rerun the corresponding steps.
from pathlib import Path
import pandas as pd
project_root = Path.cwd().parent.parent
Gold standard¶
We use the gold standard file in the data/gold_standards folder.
gold_standard_path = project_root / "data" / "gold_standards" / "exact_matching_gs.csv"
gold_standard = pd.read_csv(gold_standard_path)
gold_standard.head()
| source_concept_id | source_term | target_concept_id | target_concept_name | predicate | target_concept_id_b | target_concept_name_b | predicate_b | |
|---|---|---|---|---|---|---|---|---|
| 0 | 8690 | Unspecified enthesopathy, lower limb, excludin... | 4116324 | Enthesopathy of lower leg and ankle region | exactMatch | 4116324.0 | Enthesopathy of lower leg and ankle region | broadMatch |
| 1 | 9724 | Lead-induced chronic gout, unspecified elbow | 607432 | Chronic gout caused by lead | broadMatch | NaN | NaN | NaN |
| 2 | 9770 | Chronic gout due to renal impairment, right elbow | 46270464 | Gout of elbow due to renal impairment | broadMatch | NaN | NaN | NaN |
| 3 | 9946 | Pathological fracture, left ankle | 760649 | Pathological fracture of left ankle | exactMatch | NaN | NaN | NaN |
| 4 | 10389 | Unspecified fracture of skull | 4324690 | Fracture of skull | exactMatch | NaN | NaN | NaN |
This gold standard contains the following columns:
source_concept_id: the concept ID of the source term (non-standard concept)source_term: the source term textstandard_concept_id: the concept ID of the standard concept to which the source term mapsstandard_concept_name: the name of the standard conceptpredicate: Whether the mapping is anexactMatchor abroadMatch. OnlyexactMatchmappings are considered correct in this evaluation.target_concept_id_b,target_concept_name_b, andpredicate_b: optional second mapping for the source term that is equally valid, if available.
Term cleanup¶
The first step is to clean up the source terms, removing any extraneous information that may interfere with matching.
This includes removing phrases like "not otherwise specified", as well as other uninformative parts of the source term.
This step uses the LLM to perform the cleanup.
The TermCleaner class handles this, using the system prompts specified in the config.yaml file, and adds a cleaned_term column to the input DataFrame.
from ariadne.term_cleanup.term_cleaner import TermCleaner
cleaned_terms_file = project_root / "data" / "notebook_results" / "exact_matching_cleaned_terms.csv"
if cleaned_terms_file.exists():
cleaned_terms = pd.read_csv(cleaned_terms_file)
print("Loaded cleaned terms from file.")
else:
term_cleaner = TermCleaner()
cleaned_terms = term_cleaner.clean_terms(gold_standard)
print(f"Total LLM cost: ${term_cleaner.get_total_cost():.6f}")
cleaned_terms.to_csv(cleaned_terms_file, index=False)
cleaned_terms[["source_term", "cleaned_term"]].head(10)
Loaded cleaned terms from file.
| source_term | cleaned_term | |
|---|---|---|
| 0 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot |
| 1 | Lead-induced chronic gout, unspecified elbow | Lead-induced chronic gout, elbow |
| 2 | Chronic gout due to renal impairment, right elbow | Chronic gout due to renal impairment, right elbow |
| 3 | Pathological fracture, left ankle | Pathological fracture, left ankle |
| 4 | Unspecified fracture of skull | fracture of skull |
| 5 | Ocular laceration without prolapse or loss of ... | Ocular laceration without prolapse or loss of ... |
| 6 | Penetrating wound of orbit with or without for... | Penetrating wound of orbit |
| 7 | Fracture of unspecified shoulder girdle, part ... | Fracture of shoulder girdle |
| 8 | Drowning and submersion due to falling or jump... | Drowning and submersion |
| 9 | Alcohol use, unspecified with withdrawal delirium | Alcohol use with withdrawal delirium |
Verbatim matching¶
The next step is to perform verbatim matching, i.e. looking for exact matches of the cleaned source terms in the vocabulary. We do allow for some minor variations, such as case differences and punctuation differences.
We use the VocabVerbatimTermMapper class for this, which first needs to create an index of the vocabulary terms.
For this it will connect to the database specified in the environment variables.
It will restrict to the vocabularies and domains specified in the config.yaml file.
This adds the mapped_concept_id and mapped_concept_name columns to the input DataFrame.
from ariadne.verbatim_mapping.term_downloader import download_terms
from ariadne.verbatim_mapping.vocab_verbatim_term_mapper import VocabVerbatimTermMapper
verbatim_match_file = project_root / "data" / "notebook_results" / "exact_matching_verbatim_maps.csv"
if verbatim_match_file.exists():
verbatim_matches = pd.read_csv(verbatim_match_file)
print("Loaded verbatim matches from file.")
else:
download_terms() # Downloads the terms as Parquet files to the folder specified in config.yaml.
verbatim_mapper = VocabVerbatimTermMapper() # Will construct the vocabulary index if needed
verbatim_matches = verbatim_mapper.map_terms(cleaned_terms)
verbatim_matches.to_csv(verbatim_match_file, index=False)
verbatim_matches[["source_term", "cleaned_term", "mapped_concept_id", "mapped_concept_name"]].head(10)
Loaded verbatim matches from file.
| source_term | cleaned_term | mapped_concept_id | mapped_concept_name | |
|---|---|---|---|---|
| 0 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | -1 | NaN |
| 1 | Lead-induced chronic gout, unspecified elbow | Lead-induced chronic gout, elbow | -1 | NaN |
| 2 | Chronic gout due to renal impairment, right elbow | Chronic gout due to renal impairment, right elbow | -1 | NaN |
| 3 | Pathological fracture, left ankle | Pathological fracture, left ankle | -1 | NaN |
| 4 | Unspecified fracture of skull | fracture of skull | 4324690 | Fracture of skull |
| 5 | Ocular laceration without prolapse or loss of ... | Ocular laceration without prolapse or loss of ... | -1 | NaN |
| 6 | Penetrating wound of orbit with or without for... | Penetrating wound of orbit | 4334734 | Penetrating wound of orbit |
| 7 | Fracture of unspecified shoulder girdle, part ... | Fracture of shoulder girdle | -1 | NaN |
| 8 | Drowning and submersion due to falling or jump... | Drowning and submersion | -1 | NaN |
| 9 | Alcohol use, unspecified with withdrawal delirium | Alcohol use with withdrawal delirium | -1 | NaN |
Embedding vector search¶
Next, we perform embedding vector search for the source terms that were not matched by verbatim matching.
This uses the vector store specified in the config.yaml file, which can be either a local PGVector instance or the OHDSI Hecate API.
from ariadne.vector_search.pgvector_concept_searcher import PgvectorConceptSearcher
# from ariadne.vector_search.hecate_concept_searcher import HecateConceptSearcher
vector_search_results_file = project_root / "data" / "notebook_results" / "exact_matching_vector_search_results.csv"
if vector_search_results_file.exists():
vector_search_results = pd.read_csv(vector_search_results_file)
print("Loaded vector search results from file.")
else:
concept_searcher = PgvectorConceptSearcher()
# concept_searcher = HecateConceptSearcher()
unmatched_terms = cleaned_terms.copy()
unmatched_terms = unmatched_terms[unmatched_terms["source_concept_id"].isin(
verbatim_matches["source_concept_id"][verbatim_matches["mapped_concept_id"] == -1]
)]
vector_search_results = concept_searcher.search_terms(
unmatched_terms, term_column="cleaned_term"
)
vector_search_results.to_csv(vector_search_results_file, index=False)
vector_search_results[["source_term", "cleaned_term", "matched_concept_id", "matched_concept_name", "match_score"]].head(10)
Loaded vector search results from file.
| source_term | cleaned_term | matched_concept_id | matched_concept_name | match_score | |
|---|---|---|---|---|---|
| 0 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 4194889 | Enthesopathy of lower limb | 0.071184 |
| 1 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 46284922 | Enthesitis of lower limb | 0.233134 |
| 2 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 198846 | Enthesopathy of hip region | 0.237688 |
| 3 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 4347178 | Enthesopathy of foot region | 0.264289 |
| 4 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 73008 | Enthesopathy | 0.267237 |
| 5 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 4116324 | Enthesopathy of lower leg and ankle region | 0.270949 |
| 6 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 37108924 | Enthesopathy of left foot | 0.285147 |
| 7 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 37309727 | Bilateral enthesopathy of feet | 0.307093 |
| 8 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 37108923 | Enthesopathy of right foot | 0.308362 |
| 9 | Unspecified enthesopathy, lower limb, excludin... | enthesopathy, lower limb, excluding foot | 42537682 | Enthesopathy of upper limb | 0.322310 |
Evaluating the vector search results¶
We can now evaluate the vector search results on their own, before combining them with the verbatim matches. The results are written to a text file that contains overall statistics as well as detailed per-term results.
from ariadne.evaluation.concept_search_evaluator import evaluate_concept_search
evaluate_concept_search(
search_results=vector_search_results,
output_file=project_root / "data" / "notebook_results" / "exact_matching_vector_search_evaluation.txt",
)
with open(project_root / "data" / "notebook_results" / "exact_matching_vector_search_evaluation.txt", "r") as f:
evaluation_text = f.read()
print("\n".join(evaluation_text.split("\n")[:15]))
Evaluation complete. Results written to E:\git\Ariadne\data\notebook_results\exact_matching_vector_search_evaluation.txt
Evaluated gold standard concepts: 333
Mean Average Precision: 0.8651153955075523
Recall@1: 0.7957957957957958
Recall@3: 0.924924924924925
Recall@10: 0.978978978978979
Recall@25: 0.993993993993994
Source term: Unspecified enthesopathy, lower limb, excluding foot (8690)
Searched term: enthesopathy, lower limb, excluding foot
Gold standard concept rank: 6
match_rank Correct matched_concept_id matched_concept_name
1 4194889 Enthesopathy of lower limb
2 46284922 Enthesitis of lower limb
3 198846 Enthesopathy of hip region
LLM exact matching¶
Finally, we perform LLM-based exact matching for the source terms that were not matched by verbatim matching, by choosing from a set of candidate concepts retrieved by vector search.
Retrieving concept context¶
We observed the LLM does require some context about the target concepts, including the parents, children, and synonyms, to make good mapping decisions. We must therefore retrieve this context from the database before passing it to the LLM.
from ariadne.llm_mapping.concept_context_retriever import add_concept_context
context_file_name = project_root / "data" / "notebook_results" / "exact_matching_vector_search_context.csv"
if context_file_name.exists():
vector_search_results = pd.read_csv(context_file_name)
print("Loaded vector search context from file.")
else:
vector_search_results = add_concept_context(vector_search_results)
vector_search_results.to_csv(context_file_name, index=False)
vector_search_results[["source_term", "matched_concept_id", "matched_concept_name", "matched_synonyms", "matched_concept_class_id", "matched_children"]].head(10)
Loaded vector search context from file.
| source_term | matched_concept_id | matched_concept_name | matched_synonyms | matched_concept_class_id | matched_children | |
|---|---|---|---|---|---|---|
| 0 | Unspecified enthesopathy, lower limb, excludin... | 4194889 | Enthesopathy of lower limb | Enthesopathy of lower limb (disorder) | Disorder | Enthesopathy of hip region;Enthesopathy of ank... |
| 1 | Unspecified enthesopathy, lower limb, excludin... | 46284922 | Enthesitis of lower limb | Enthesitis of lower limb (disorder) | Disorder | Enthesitis of knee;Enthesitis of quadriceps te... |
| 2 | Unspecified enthesopathy, lower limb, excludin... | 198846 | Enthesopathy of hip region | Enthesopathy of hip region (disorder) | Disorder | Enthesopathy of left hip;Enthesopathy of right... |
| 3 | Unspecified enthesopathy, lower limb, excludin... | 4347178 | Enthesopathy of foot region | Enthesopathy of foot region (disorder) | Disorder | Tarsus enthesopathy;Enthesopathy of right foot... |
| 4 | Unspecified enthesopathy, lower limb, excludin... | 73008 | Enthesopathy | Enthesopathy (disorder) | Disorder | Peripheral enthesopathy;Enthesitis;Spinal enth... |
| 5 | Unspecified enthesopathy, lower limb, excludin... | 4116324 | Enthesopathy of lower leg and ankle region | Enthesopathy of lower leg and ankle region (di... | Disorder | NaN |
| 6 | Unspecified enthesopathy, lower limb, excludin... | 37108924 | Enthesopathy of left foot | Enthesopathy of left foot (disorder) | Disorder | Bilateral enthesopathy of feet;Enthesopathy of... |
| 7 | Unspecified enthesopathy, lower limb, excludin... | 37309727 | Bilateral enthesopathy of feet | Enthesopathy of bilateral feet (disorder);Enth... | Disorder | NaN |
| 8 | Unspecified enthesopathy, lower limb, excludin... | 37108923 | Enthesopathy of right foot | Enthesopathy of right foot (disorder) | Disorder | Bilateral enthesopathy of feet;Enthesopathy of... |
| 9 | Unspecified enthesopathy, lower limb, excludin... | 42537682 | Enthesopathy of upper limb | Enthesopathy of upper limb (disorder) | Disorder | Enthesopathy of elbow region;Enthesopathy of s... |
LLM mapping¶
Now we can perform the LLM-based exact matching.
This uses the system prompts specified in the config.yaml file, which are tailored for GPT o3.
LLM responses are cached to avoid duplicate costs when rerunning.
from ariadne.llm_mapping.llm_mapper import LlmMapper
llm_mapper = LlmMapper()
mapped_terms = llm_mapper.map_terms(vector_search_results)
llm_mapped_terms_file = project_root / "data" / "notebook_results" / "exact_matching_llm_mapped_terms.csv"
mapped_terms.to_csv(llm_mapped_terms_file, index=False)
print(f"Total LLM cost: ${llm_mapper.get_total_cost():.6f}")
mapped_terms[["source_term", "cleaned_term", "mapped_concept_id", "mapped_concept_name"]].head(10)
Total LLM cost: $0.000000
| source_term | cleaned_term | mapped_concept_id | mapped_concept_name | |
|---|---|---|---|---|
| 0 | AV block first degree | AV block first degree | 314379 | First degree atrioventricular block |
| 1 | Abnormal findings in cerebrospinal fluid | Abnormal findings in cerebrospinal fluid | 4065770 | Cerebrospinal fluid examination abnormal |
| 2 | Abnormal findings in cerebrospinal fluid, Abno... | Abnormal findings in cerebrospinal fluid, Abno... | 4172064 | Abnormal histological findings in CSF |
| 3 | Abnormal microbiological findings in specimens... | Abnormal microbiological findings in specimens... | 4171911 | Abnormal microbiological finding in specimen f... |
| 4 | Surgical operation with formation of external ... | Abnormal reaction or later complication due to... | 4123283 | Disorder of stoma |
| 5 | Abnormalities of heart beat | Abnormalities of heart beat | 4262562 | Abnormal heart beat |
| 6 | Abscess of breast associated with pregnancy, u... | Abscess of breast associated with pregnancy | 78218 | Obstetric breast abscess with antenatal compli... |
| 7 | Abscess of bursa, right ankle and foot | Abscess of bursa, right ankle and foot | -1 | no_match |
| 8 | Accidents caused by transmission machinery | Accidents caused by transmission machinery | 439456 | Accident caused by power transmission machinery |
| 9 | Acn� excori�e | Acn� excori�e | 4004094 | Excoriated acne |
Final evaluation¶
Now we can combine the verbatim matches and the LLM-based matches, and evaluate the overall performance.
from ariadne.evaluation.concept_selection_evaluator import evaluate
# Combine verbatim matches and LLM matches
# First take all verbatim matches that were successful
final_mapped_terms = verbatim_matches[verbatim_matches["mapped_concept_id"] != -1][
["source_concept_id", "source_term", "cleaned_term", "mapped_concept_id", "mapped_concept_name"]
].copy()
final_mapped_terms["map_method"] = "verbatim"
# Then add the LLM matches (for the terms that were not verbatim matched)
llm_mapped_terms_filtered = mapped_terms[
["source_concept_id", "source_term", "cleaned_term", "mapped_concept_id", "mapped_concept_name", "mapped_rationale"]
].copy()
llm_mapped_terms_filtered["map_method"] = "llm"
final_mapped_terms = pd.concat([final_mapped_terms, llm_mapped_terms_filtered], ignore_index=True)
# Evaluate
final_evaluation_results = evaluate(final_mapped_terms)
final_evaluation_results.to_csv(project_root / "data" / "notebook_results" / "exact_matching_final_evaluation.csv", index=False)
final_evaluation_results[["source_term", "target_concept_name", "map_method", "mapped_concept_name", "is_correct"]].head()
| source_term | target_concept_name | map_method | mapped_concept_name | is_correct | |
|---|---|---|---|---|---|
| 0 | Unspecified fracture of skull | Fracture of skull | verbatim | Fracture of skull | True |
| 1 | Penetrating wound of orbit with or without for... | Penetrating wound of orbit | verbatim | Penetrating wound of orbit | True |
| 2 | Other retinal disorders in diseases classified... | Retinal disorder | verbatim | Retinal disorder | True |
| 3 | Other forms of angina pectoris | Angina pectoris | verbatim | Angina pectoris | False |
| 4 | Other forms of acute ischemic heart disease | Acute ischemic heart disease | verbatim | Acute ischemic heart disease | True |
overall_accuracy = final_evaluation_results["is_correct"].mean()
print(f"Overall accuracy: {overall_accuracy:.4f}")
Overall accuracy: 0.8836
# Zip data/llm_responses folder to data/notebook_results/llm_responses.zip
import shutil
shutil.make_archive(
project_root / "data" / "notebook_results" / "llm_responses",
'zip',
project_root / "data" / "llm_mapper_responses"
)
print("Zipped LLM responses.")
Zipped LLM responses.