Chapter Leads: Anna Ostropolets
This is page is currently a stub. The chapter is being written in the OHDSI Teams directory. When the draft is complete, it will be converted to markdown and moved to this file.
Author Resources (requires an OHDSI Teams account):
Public Resources:
- Book of OHDSI, Edition 1
- Source Code for Book of OHDSI, Edition 1
- OHDSI Home Page
The OHDSI Standardized Vocabularies are a foundational part of the OHDSI research network, and an integral part of the Common Data Model (CDM) (1). They allow standardization of methods, definitions, and results by defining the content of the data, paving the way for true remote (behind the firewall) network research and analytics. Usually, finding and interpreting the content of observational healthcare data, whether it is structured data using coding schemes or laid down in free text, is passed all the way through to the researcher, who is faced with a myriad of different ways to describe clinical events. OHDSI requires harmonization not only to a standardized format, but also to a rigorous standard content.
The Vocabularies are a collection of public standard vocabularies and terminologies used in the network, which we consolidate from their different original formats and life-cycle conventions into the CDM table structure. The system is dynamic, it evolves with frequent source vocabulary updates, deprecations, and concept replacements, all of which are version-controlled and available via ATHENA - OHDSI’s vocabulary distribution platform (2). Vocabularies support phenotyping, covariate construction, large-scale analytics and result reporting and is a product of community effort: Vocabulary Team maintains and improves vocabularies according to the roadmap (3), vocabulary stewards maintain individual vocabularies (4), various Workgroups coordinate efforts for special use cases such as device or vaccine harmonization and contributors across the community add their vocabulary content and improve existing, particularly through enhancing mappings (5).
In this chapter we first describe the main principles of the Standardized Vocabularies and processes. We will walk through Vocabularies components and some typical situations, all of which are necessary to understand and utilize this foundational resource. We also point out where the support of the community is required to continuously improve it and describe special situations that require further development.
5.1 Why Vocabularies, and Why Standardizing
Medical vocabularies go back to the Bills of Mortality in medieval London to manage outbreaks of the plague and other diseases (Figure 5.1).
Since then, the classifications have greatly expanded in size and complexity and spread into other aspects of healthcare, such as procedures and services, drugs, medical devices, etc. The main principles have remained the same: they are controlled vocabularies, terminologies, hierarchies, or ontologies that some healthcare communities agree upon for the purpose of capturing, classifying, and analyzing patient data. Many of these vocabularies are maintained by public and government agencies with a long-term mandate for doing so. For example, the World Health Organization (WHO) produces the International Classification of Disease (ICD) with the recent addition of its 11th revision (ICD11). Local governments create country-specific versions, such as ICD10CM (USA), ICD10GM (Germany), etc. Governments also control the marketing and sale of drugs and maintain national repositories of such certified drugs. Vocabularies are also used in the private sector, either as commercial products or for internal use, such as electronic health record (EHR) systems or for medical insurance claim reporting.
As a result, each country, region, healthcare system and institution tend to have their own classifications that would most likely only be relevant where it is used. This myriad of vocabularies prevents interoperability of the systems they are used in. Standardization is the key that enables patient data exchange, unlocks health data analysis on a global level, and allows systematic and standardized research, including performance characterization and quality assessment. To address the interoperability problem, multinational organizations have sprung up and started creating broad standards, such as the Standard Nomenclature of Medicine (SNOMED) and Logical Observation Identifiers Names and Codes (LOINC). In the US, the Health IT Standards Committee (HITAC) recommends the use of SNOMED, LOINC, and the drug vocabulary RxNorm as standards to the National Coordinator for Health IT (ONC) for use in a common platform for nationwide health information exchange across diverse entities.
OHDSI developed the OMOP CDM, a global standard for observational research. As part of the CDM, the OHDSI Standardized Vocabularies are available for two main purposes:
Common repository of all vocabularies used in the community
Standardization and mapping for use in research
The Standardized Vocabularies are available to the community free of charge and must be used for OMOP CDM instance as its mandatory reference table. It is crucial to use the most recent version of the Vocabularies and continuously incorporate new versions in the ETL as Vocabularies changes and shifts impact common research tasks (6).
5.1.1 Vocabularies Use Cases and Users
OHDSI Vocabularies are different from other ontology systems, such as the Unified Medical Language System (UMLS) (7) and the difference stems from the main use case of evidence generation that OHDSI supports. Both UMLS and OHDSI aggregate relationships from source vocabularies. UMLS provides crosswalks among vocabularies with various degrees of fidelity and such crosswalks can be incomplete or ambiguous. OHDSI curates mappings and selects high-quality ones for the official “Maps to” relationships from source vocabularies to a single reference standard, ensuring that all data sources speak the same language. For example, “Atrial fibrillation” from ICD-9, Read, MeSH, etc., are all mapped to a single SNOMED concept in the Condition domain. UMLS, in contrast, groups synonymous terms under a CUI but does not designate one as “the code to use” serving as a translation table and not enforcing a single vocabulary for data encoding. UMLS contains many international vocabularies, but historically it has had a strong U.S. focus, and some content can lag in updates. OHDSI Vocabularies explicitly integrate both US and non-US coding systems and even create new standard concepts for non-US use cases to achieve global coverage. For example, US drugs are covered in RxNorm that we import, international drugs are covered in RxNorm Extension that we create de-novo and both of them are integrated with Anatomic Therapeutic Classification (ATC) (8,9). OHDSI Vocabularies are optimized for standardized analytics, offering open-access, harmonized coverage of both U.S. and international terminologies to enable consistent, reproducible studies across institutions and countries.
Vocabularies are centered around generating real-world evidence from observational studies and are mostly used for two groups of tasks: ETL of data to OMOP CDM and subsequent research on converted data. If you are a data engineer/ETL developer, the most relevant information is how to use correct source-to-standard mappings and populate both standard and source concept ID fields appropriately. Additionally, you need to know how to track vocabulary changes adopt ETL accordingly. If you are a researcher, the most relevant information is how to use vocabularies to find relevant codes for concept sets and features, use hierarchies, and examine mappings.
5.1.2 Access to the Standardized Vocabularies
The OHDSI Standardized Vocabularies are distributed via ATHENA (2), a web-based platform for browsing and downloading vocabulary data. You can use it to search, explore, and filter vocabularies by domain, concept class, vocabulary source, standard status and validity. You can select relevant vocabularies and download a pre-packaged vocabulary bundle, ready for loading into a local OMOP CDM instance.
To download a zip file with all Standardized Vocabularies tables, select all the vocabularies you need for your OMOP CDM. Vocabularies with Standard Concepts and very common usage are preselected. Add vocabularies that are used in your source data. Vocabularies that are proprietary have no select button. Click on the “License required” button to incorporate a licensed-required vocabulary into your list. The Vocabulary Team will contact you and request you demonstrate your license or help you connect to the right body to obtain one.
Each vocabulary download includes a ZIP file containing a standard set of CSV files, which can be loaded into your database using standard SQL scripts or programmatically. You will also need to re-constitute names of CPT4 codes as per our use agreement (10).
The VOCABULARY.csv file contains the version and release date metadata for each vocabulary, which should be recorded to ensure reproducibility in analyses and network studies. When updating to a newer vocabulary version, we recommend reviewing the changes in concept definitions, domain assignments, mappings, and deprecated concepts to ensure that downstream data and cohort definitions remain valid (6).
You can also select a specific vocabulary release different from the current release or download a file that contains the delta between two given releases.
5.2 Vocabularies Process and Governance
5.2.1 Building the Standardized Vocabularies and Vocabularies Principles
All vocabularies of the Standardized Vocabularies are consolidated into the same common format: CONCEPT, CONCEPT_RELATIONSHIP, CONCEPT_ANCESTOR, CONCEPT_SYNONYM, and supporting reference files such as VOCABULARY, DOMAIN, CONCEPT_CLASS, and RELATIONSHIP. This relieves the researchers from having to understand and handle multiple different formats and life-cycle conventions of the originating vocabularies.
OHDSI generally prefers adopting existing vocabularies, rather than de-novo construction, because (i) many vocabularies have already been utilized in observational data in the community, and (ii) construction and maintenance of vocabularies is complex and requires the input of many stakeholders over long periods of time to mature. For this reason, dedicated organizations provide vocabularies, which undergo a life cycle of generation, deprecation, merging, and splitting. Currently, OHDSI only produces internal administrative vocabularies like Type Concepts (for example, condition type concepts) as well as several other vocabularies to cover areas with existing gaps: RxNorm Extension to cover drugs that are only used outside the United States, OMOP Investigational Drugs for investigational drugs, Cancer Modifier for cancer measurements, and OMOP Extension for miscellaneous gaps. There are other community-driven efforts, such as GIS Vocabulary Package (11).
All vocabularies go through several common stages upon refresh: staging or harmonization to a common table structure, normalization and creation of crosswalks, integration with other vocabularies and release (12). All steps are accompanied by a set of quality assurance and control procedures, both automated and human-curated (13).
OHDSI Vocabularies follow twelve principles (14). Among others, Vocabularies focus on and support OHDSI use case of generating new evidence. They meant to be comprehensive, that is there are enough concepts to cover any event relevant to the patient’s healthcare experience (e.g., conditions, procedures, exposures to drug, etc.) as well as some of the administrative information of the healthcare system (e.g., visits, care sites, etc.). They strive to have unique standard concept, where for each Clinical Entity there is only one concept representing it, called the Standard Concept. Other equivalent or similar concepts are designated non-Standard and mapped to the Standard ones. Moreover, such concepts should be stated as fact, no negations of facts, no reference to the past, and no flavors of null (unknown, not reported, etc.).
5.2.2 Vocabularies Governance, Roadmap and Role of the Community
OHDSI Vocabularies work and processes are governed by the OHDSI Central Coordinating Center’s body, Vocabulary Committee, which includes representatives from across the OHDSI community and helps set priorities for maintenance, content expansion, and quality improvement. Committee’s work is based on the landscape assessment conducted in 2023 (15). As a part of this work, it approves the roadmap for bi-annual releases of the Vocabularies.
Release happens in February and August and is accompanied by detailed roadmap updates and release notes describing the changes (16). Each release note describes (1) newly added vocabularies, (2) concepts and/or mappings newly added to the existing vocabularies, (4) changes in mappings, domains, status of the concepts as well as detailed description of the actions performed for specific vocabularies. Additionally, the release notes contain the artifacts not available through Athena: pack content, SSSOM-compatible metadata (17) for concepts and relationships as well as reports for alignment with vocabulary principles (August 2024 release).
You can use information about releases and the roadmap in two ways. First, you can assess the content of each release and use open source tools to assess its impact on ETL and research (6,18). If you are a ETLer or are responsible for your institution’s data/OMOP CDM, you should update OMOP CDM instance with the latest version of the Vocabularies to benefit from improved coverage, consistency, and corrected mappings. Vocabularies change a lot. If you are not updating, you are falling behind in research.
Second, you can use this information to assess if planned activities meet your needs. You assist in vocabulary maintenance if your vocabulary is not on the roadmap as a vocabulary steward (19). With each release stewards from the community refresh and improve their vocabularies, even if they are not on the roadmap (current list of stewards can be found here (4)). You can also add your vocabulary, concepts or improve existing content (mappings, domains) through community contribution (5).
5.2.2.1 Community contributions
The extensive scope of the OMOP Standard Vocabularies poses a challenge to maintenance and scalability. The OHDSI Vocabulary Team focuses on core terminologies out of necessity. However, there are plenty of opportunities for the OHDSI community to assist in vocabulary maintenance. Examples above regarding improvements in mappings, labels, synonyms, etc. are welcomed from the community. Working Groups may feel particular ownership of a domain or specialty area and wish to help manage the necessary vocabulary. When this happens, the core terminology management system can be extended through integration of community-contributed content. Begun in earnest in 2024, this community and centrally-management vocabulary integration allows for more scalable contribution and more rapid conceptual gap-filling. A community-contribution infrastructure has been developed in phases depending on the complexity of the contribution. Small changes or additions can be provided using templates.
You can use templates to add a new standard vocabulary, add non-standard concepts, add mappings, change mappings or concept domains, or propose upgrading a non-standard concept to standard. Modification of content requires community approval through the Vocabulary Workgroup. Template submissions should be completed and ratified two months prior to the release date (end of June/end of December for August and February releases, respectively). Instructions for completed templates are described on the GitHub Wiki (5).
Larger contributions (for example, entire terminologies or drug catalogues) require staging and integration using a compatible environment to that used for managing the core terminologies. Examples of community-staged contributions include the Heme-Onc vocabulary, the Veterinary vocabulary and the CIEL terminology. For complex contributions, it is best to have a working group sponsor your request. You can use the instructions provided on Wiki under Community Contributions Part II (5). We recommend you talk to the members of the Vocabulary Workgroup or Team to discuss your specific use case.
5.3 OHDSI Vocabularies Structure: Concepts and Relationships
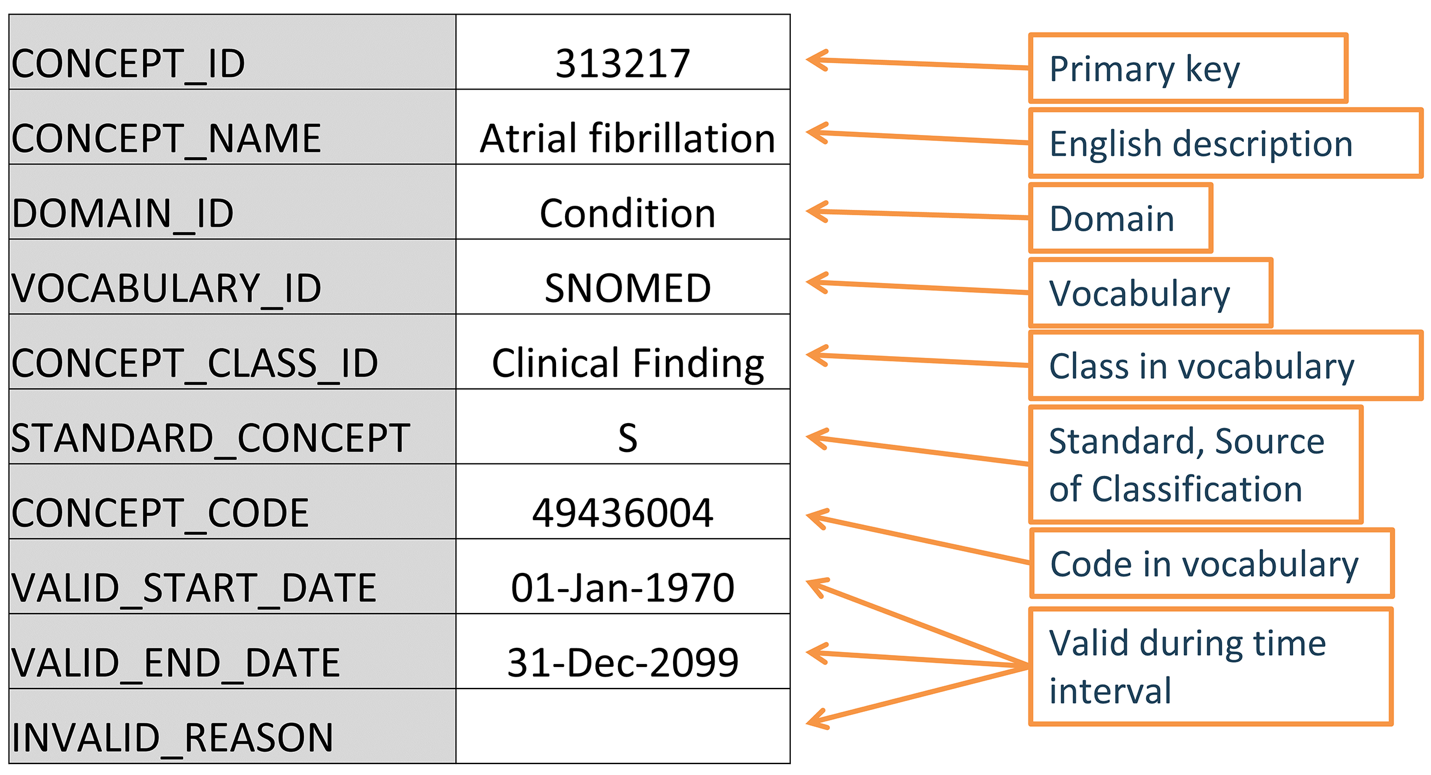
All clinical events in the OMOP CDM are represented as concepts, which capture the semantic notion of each event. They are the fundamental building blocks of the data records, making almost all tables fully normalized with few exceptions. Concepts are stored in the CONCEPT table (Figure 5.2).
CONCEPT table record for the SNOMED code for Atrial Fibrillation.
5.3.1 Concept IDs
Each concept is assigned a concept ID to be used as a primary key. This meaningless integer ID, rather than the original code from the source vocabulary, is used to record data in the CDM event tables via the foreign key fields. No two concepts (even from different vocabularies) share the same ID. Conversely, the same source code might appear in multiple vocabularies, but each distinct concept gets its own ID.
5.3.2 Concept Names
Each concept has one name. Names are always in English. They are imported from the source of the vocabulary. If the source vocabulary has more than one name, the most expressive (fully specified) is selected and the remaining ones are stored in the CONCEPT_SYNONYM table under the same CONCEPT_ID key. Non-English names are recorded in CONCEPT_SYNONYM as well, with the appropriate language concept ID in the LANGUAGE_CONCEPT_ID field. The name can only be 255 characters long, which means that very long names get truncated, and the full-length version recorded as another synonym, which can hold up to 1000 characters. Tools like Athena and ATLAS use the concept names and synonyms to let users search for concepts. When doing analysis, it is often convenient to have the concept names for interpretability, but analysis logic should use the CONCEPT_ID.
5.3.3 Domains
Each concept is assigned a domain in the DOMAIN_ID field, which, in contrast to the numerical CONCEPT_ID, is a short, case-sensitive, unique alphanumeric ID for the domain. Domains are OMOP-specific and correspond to the OMOP CDM tables (20). Examples of such identifiers are “Condition,” “Drug,” “Procedure,” “Visit,” “Device,” “Specimen,” etc. Domains also direct to which CDM table and field a clinical event or event attribute is recorded. For example, “Atrial fibrillation” is a clinical finding that would be recorded in the Condition Occurrence table, so its domain is “Condition”; a concept for a lab test (for example, “Blood glucose measurement”) would have domain “Measurement” and belong in the Measurement table. Domains are assigned to codes, and a vocabulary can have different domains: for example, HCPCS, while considered procedure vocabulary, also has codes with Drug and Observation domains.
The domain heuristic follows the definitions of the domains. These definitions are derived from the table and field definitions in the CDM {Chapter 4}. The heuristic is not perfect; there are grey zones ({Section 5.4} ”Special Situations”), source vocabulary shifts, and occasional misassignments. Although domains of concepts may change, 95% of the concepts never changed their domain since Vocabularies’ inception (for more information, see Assets in v20240830 release notes) (16).
5.3.4 Vocabularies
Each vocabulary has a short case-sensitive unique alphanumeric ID, which generally follows the abbreviated name of the vocabulary, omitting dashes. For example, ICD-9-CM has the vocabulary ID “ICD9CM”. As of 2025, over 140 vocabularies are available through ATHENA and follow different cadence of updates. The source and the version of the vocabularies is defined in the VOCABULARY reference file and documentation for individual vocabularies can be found on GitHub (4,16).
5.3.5 Concept Classes
Some vocabularies classify their codes or concepts, denoted through their case-sensitive unique alphanumerical IDs. For example, SNOMED has 33 such concept classes, which SNOMED refers to as “semantic tags”: clinical finding, social context, body structure, etc. These are vertical divisions of the concepts. Others, such as MedDRA or RxNorm, have concept classes classifying horizontal levels in their stratified hierarchies. Vocabularies without any concept classes, such as HCPCS, use the vocabulary ID as the Concept Class ID.
| Concept class subdivision principle | Vocabulary |
|---|---|
| Horizontal | All drug vocabularies, CDТ, Episode, HCPCS, HemOnc, ICDs, MedDRA, OSM, Census |
| Vertical | CIEL, HES Specialty, ICDO3, MeSH, NAACCR, NDFRT, OPCS4, PCORNET, Plan, PPI, Provider, SNOMED, SPL, UCUM |
| Mixed | CPT4, ISBT, LOINC |
| None | OXMIS, Race, Revenue Code, Sponsor, Supplier, UB04s, Visit |
Horizontal concept classes allow you to determine a specific hierarchical level. For example, in the drug vocabulary RxNorm, the concept class “Ingredient” defines the top level of the hierarchy. In the vertical model, members of a concept class can be of any hierarchical level from the top to the very bottom. Concept class is mostly a descriptive attribute and helps to filter concepts. For example, if you only want to select drugs with a specific Brand Name you can filter to “Branded Drug” class.
5.3.6 Standard Concepts
A Standard Concept is the community-endorsed, canonical representation of a clinical meaning within the OHDSI Vocabularies. It serves as the unified semantic identifier for a specific entity (for example, condition, drug, procedure), regardless of how that entity is expressed in source vocabularies. Only Standard Concepts are used to populate the CONCEPT_ID fields in the CDM, ensuring consistency across diverse datasets. Standard concepts serve as the target for mappings. For each clinical entity, one concept from one vocabulary is chosen to be standard. This becomes the “hub” to which all equivalent source codes are mapped. For example, MESH code D001281, CIEL code 148203, SNOMED code 49436004, ICD9CM code 427.31 and Read code G573000 all define “Atrial fibrillation” in the condition domain, but only the SNOMED concept is Standard and represents the condition in the data. The others are designated non-standard or source concepts and mapped to the Standard ones. Standard Concepts are indicated through an “S” in the STANDARD_CONCEPT field. And only these Standard Concepts are used to record data in the CDM fields ending in _CONCEPT_ID.
We rely on well-known reference terminologies for standard terms: SNOMED CT for conditions, RxNorm and RxNorm Extension for drugs, LOINC and SNOMED for measurements, etc. Not all concepts in those vocabularies are necessarily standard. Occasionally, a concept in a standard vocabulary might be deemed out of scope or duplicative and not used. Conversely, some concepts from typically non-standard vocabularies might be made standard if no better alternative exists.
While we strive to align with the unique standard concept principle to have one Standard concept per semantic entity, duplicates exist. For example, no deduplication of standard concepts has been performed for the Device domain. While concept mappings avoid direct collisions, this dual-standard condition can introduce ambiguity in cohort definition, concept set construction, and analytic interpretation. This phenomenon is not a flaw but rather a reflection of ontology convergence in progress, where two high-quality terminologies independently arrive at comparable representations of the same clinical reality. OHDSI addresses these cases through community review, classification logic, and long-term efforts toward consolidation via concept deprecation, reclassification, or updated mappings. Until then, such duplications must be handled with care in concept set design and ETL strategies.
5.3.7 Non-Standard Concepts
Non-standard concepts are not used in standardized analytics, but they are still part of the Standardized Vocabularies and are often found in the source data. For that reason, they are also called “source concepts”. The conversion of source concepts to Standard Concepts is a process called “mapping”.
Some of the non-standard concepts cannot be mapped and are not suitable for analytic use. Examples of such include terms like “Not reported”, “Not specified”, “Passport number” and more.
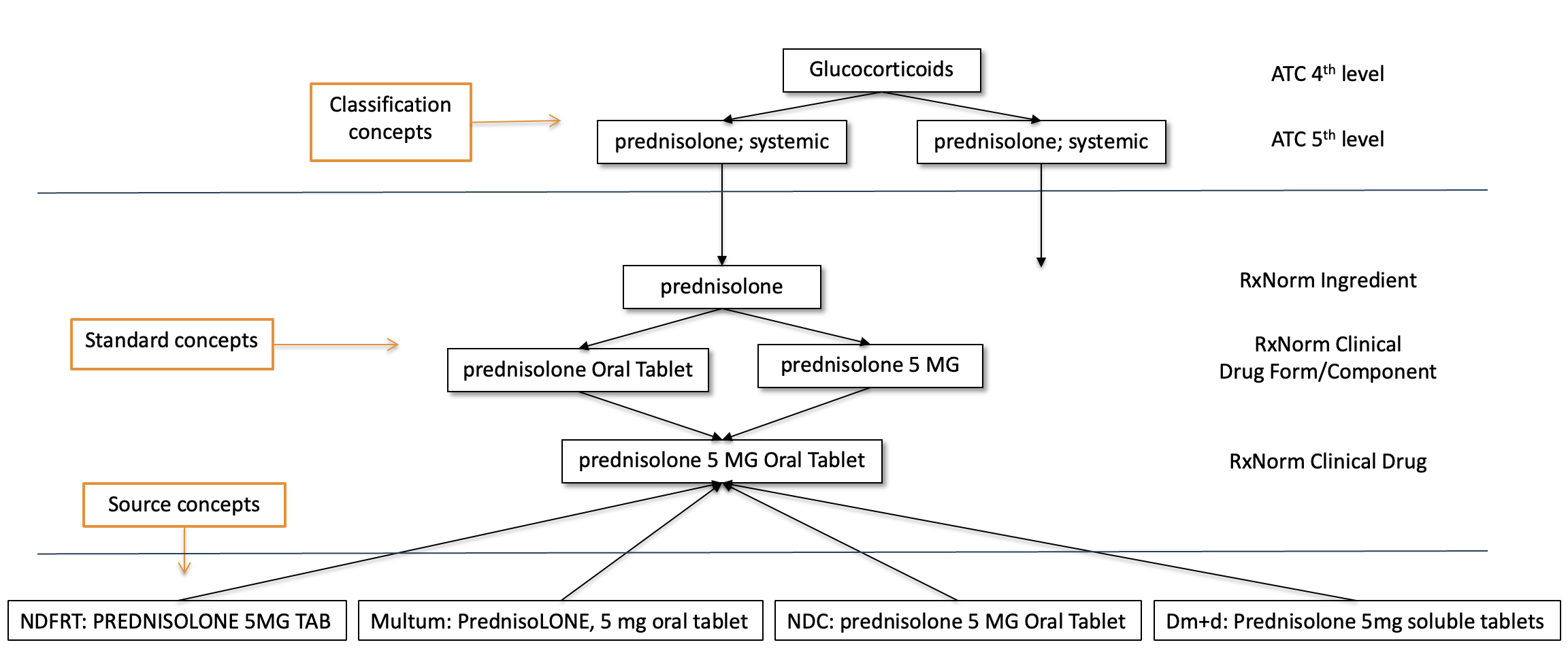
5.3.8 Classification Concepts
These concepts are not Standard and hence cannot be used to represent the data. But they are participating in the hierarchy with the Standard Concepts and can therefore be used to perform hierarchical queries. For example, querying for all descendants of ATC code prednisolone;systemic will retrieve the Standard RxNorm concept for prednisolone 5 MG Oral Tablet (Figure 5.3).
Classification concepts are marked with a “C” in the STANDARD_CONCEPT field. Most classification concepts form a hierarchy along with the standard concepts, and these relationships are stored in the CONCEPT_ANCESTOR table.
Classification concepts are vital in enabling concept set expansion via ancestry traversal. While they cannot be used to populate clinical event tables directly, they serve as entry points into clinically meaningful groupings (for example, drug classes or disorder categories). They are especially powerful when used in cohort definitions or phenotype algorithms that require aggregation of clinically related Standard Concepts. For example, selecting the ATC classification concept C09AA ACE inhibitors will retrieve all Standard RxNorm ingredients and products mapped as descendants.
Quality and coverage of classification hierarchies vary across domains. Currently, Drug and Condition domains have mature classification structures (for example, ATC, MedDRA), while Procedure, Measurement, and Device domains lack formal classification vocabularies. Caution should be exercised when interpreting classification-derived hierarchies, as they may not always reflect clinical practice or data granularity.
5.2.8.1 Standard/non-standard/classification Concept Designation
The choice of concept designation as Standard, non-standard, and classification is typically done for each domain separately at the vocabulary level. This is based on the quality of the concepts, the built-in hierarchy, and the declared purpose of the vocabulary. Also, not all concepts in a vocabulary are used as Standard Concepts. The designation is separate for each domain, each concept must be active, and there might be an order of precedence if more than one concept from different vocabularies compete for the same meaning. See Table 5.2 for examples.
| Domain | for Standard Concepts | for source concepts | for classification concepts |
|---|---|---|---|
| Condition | SNOMED, ICDO3 | SNOMED Veterinary | MedDRA |
| Procedure | SNOMED, CPT4, HCPCS, ICD10PCS, ICD9Proc, OPCS4 | SNOMED Veterinary, HemOnc, NAACCR | None at this point |
| Measurement | SNOMED, LOINC | SNOMED Veterinary, NAACCR, CPT4, HCPCS, OPCS4, PPI | None at this point |
| Drug | RxNorm, RxNorm Extension, CVX | HCPCS, CPT4, HemOnc, NAACCR | ATC |
| Device | SNOMED | Others, currently not normalized | None at this point |
| Observation | SNOMED | Others | None at this point |
| Visit | CMS Place of Service, ABMT, NUCC | SNOMED, HCPCS, CPT4, UB04 | None at this point |
5.3.9 Concept Codes
Concept codes are the identifiers used in the source vocabularies. For example, ICD9CM or NDC codes are stored in this field, while the OMOP tables use the concept ID as a foreign key into the CONCEPT table. The reason is that the name space overlaps across vocabularies, that is the same code can exist in different vocabularies with completely different meanings (Table 5.3).
| Concept ID | Concept Code | Concept Name |
Domain ID |
Vocabulary ID |
Concept Class |
|---|---|---|---|---|---|
| 35803438 | 1001 | Granulocyte colony-stimulating factors | Drug | HemOnc | Component Class |
| 35942070 | 1001 | AJCC TNM Clin T | Measurement | NAACCR | NAACCR Variable |
| 1036059 | 1001 | Antipyrine | Drug | RxNorm | Ingredient |
| 38003544 | 1001 | Residential Treatment - Psychiatric | Revenue Code | Revenue Code | Revenue Code |
| 43228317 | 1001 | Aceprometazine maleate | Drug | BDPM | Ingredient |
| 45417187 | 1001 | Brompheniramine Maleate, 10 mg/mL injectable solution | Drug | Multum | Multum |
| 45912144 | 1001 | Serum | Specimen | CIEL | Specimen |
CONCEPT_CODE is unique only within a given vocabulary. You should not join datasets via CONCEPT_CODE unless constrained by VOCABULARY_ID.
In addition, certain vocabularies, such as HCPCS, NDC, and DRG are known to reuse codes over time, assigning new meanings to previously used codes. In such cases, Vocabularies differentiate concepts based on validity dates (VALID_START_DATE, VALID_END_DATE) and keep the most recent meaning.
Some OMOP-specific vocabularies (for example, Type Concept, Visit) contain system-generated concept codes rather than real-world codes. Finally, certain source vocabularies (such as ATC or hierarchical clinical classifications) embed structural hierarchy into their codes (ATC G03E vs. G03EK), meaning that not all CONCEPT_CODE matches imply equivalence at the clinical level.
5.3.10 Lifecycle
Vocabularies are rarely permanent corpora with a fixed set of codes. Instead, codes and concepts are added and get deprecated. The OMOP CDM is a model to support longitudinal patient data, which means it needs to support concepts that were used in the past and might no longer be active, as well as supporting new concepts and placing them into context. There are three fields in the CONCEPT table that describe the possible life-cycle statuses: VALID_START_DATE, VALID_END_DATE, and INVALID_REASON. Their values differ depending on the concept life-cycle status:
Active or new concept
Description: Concept in use.
VALID_START_DATE: Day of instantiation of concept; if that is not known, day of incorporation of concept in Vocabularies; if that is not known, 1970-1-1.VALID_END_DATE: Set to 2099-12-31 as a convention to indicate “Might become invalid in an undefined future, but active right now”.INVALID_REASON: NULL
Deprecated Concept with no successor
Description: Concept inactive and cannot be used as Standard.
VALID_START_DATE: Day of instantiation of concept; if that is not known, day of incorporation of concept in Vocabularies; if that is not known, 1970-1-1.VALID_END_DATE: Day in the past indicating deprecation, or if that is not known, day of vocabulary refresh where concept in vocabulary went missing or set to inactive.INVALID_REASON: “D”
Upgraded Concept with successor
Description: Concept inactive but has defined successor. These are typically concepts which went through de-duplication.
VALID_START_DATE: Day of instantiation of concept; if that is not known, day of incorporation of concept in Vocabularies; if that is not known, 1970-1-1.VALID_END_DATE: Day in the past indicating an upgrade, or if that is not known day of vocabulary refresh where the upgrade was included.INVALID_REASON: “U”
Reused code for another new concept
Description: The vocabulary reused the concept code of this deprecated concept for a new concept.
VALID_START_DATE: Day of instantiation of concept; if that is not known, day of incorporation of concept in Vocabularies; if that is not known, 1970-1-1.VALID_END_DATE: Day in the past indicating deprecation, or if that is not known day of vocabulary refresh where concept in vocabulary went missing or set to inactive.
In addition to concept lifecycle management, the CONCEPT_RELATIONSHIP table also has lifecycle fields (VALID_START_DATE, VALID_END_DATE, INVALID_REASON) for relationships. Relationships may change over time independently of the concepts themselves. While all relationships are versioned in the internal vocabulary system, only active mappings are included in Athena downloads. Every OMOP CDM instance should record the vocabulary version (stored in the VOCABULARY table) used at ETL time to ensure transparency and reproducibility. Lifecycle management principles apply equally to custom extensions and community-contributed vocabularies: all new concepts and mappings must carry valid VALID_START_DATE entries and, when deprecated, clearly marked VALID_END_DATE and INVALID_REASON values.
5.3.11 Relationships
Any two concepts can have a defined relationship, regardless of whether the two concepts belong to the same domain or vocabulary. The nature of the relationships is indicated in its short case-sensitive unique alphanumeric ID in the RELATIONSHIP_ID field of the CONCEPT_RELATIONSHIP table. Relationships are symmetrical, that is for each relationship an equivalent relationship exists, where the content of the fields CONCEPT_ID_1 and CONCEPT_ID_2 are swapped, and the RELATIONHSIP_ID is changed to its opposite. For example, the “Maps to” relationship has an opposite relationship “Mapped from.” Different types of relationships serve different analytic purposes. “Maps to” and “Mapped from” support source-to-standard mappings. “Is a” and “Subsumes” define hierarchical subclass relationships. “Has ingredient” and “Ingredient of” structure drug compositions. “Concept replaced by” and “Concept replaces” handle lifecycle transitions across deprecated content.
As stated in the previous section, CONCEPT_RELATIONSHIP table records also have life-cycle fields VALID_START_DATE, VALID_END_DATE and INVALID_REASON. However, only active records with INVALID_REASON = NULL are available through ATHENA. Inactive relationships are kept for internal processing only.
The RELATIONSHIP table serves as the reference with the full list of relationship IDs and their reverse counterparts. It also specifies two important flags: DEFINES_ANCESTRY, indicating whether a relationship should contribute to the CONCEPT_ANCESTOR table, and IS_HIERARCHICAL, indicating whether the relationship encodes a subsumption hierarchy. Not all relationships define ancestry; only those intended to build domain hierarchies (for example, “Is a”) are used to populate CONCEPT_ANCESTOR. It is essential to distinguish between direct relationships (stored in CONCEPT_RELATIONSHIP) and inferred multi-level hierarchies (precomputed and stored in CONCEPT_ANCESTOR), especially when writing concept set queries, building phenotypes, or exploring ontology structures.
5.3.12 Mapping Relationships
These relationships provide translations from non-standard to Standard concepts, supported by two relationship ID pairs (Table 5.4).
| Relationship ID pair |
Purpose |
|---|---|
| “Maps to” and “Mapped from” |
Mapping to Standard Concepts. Standard Concepts are mapped to themselves, non-standard concepts to Standard Concepts. Most non-standard and all Standard Concepts have this relationship to a Standard Concept. The former are stored in *_SOURCE_CONCEPT_ID, and the latter in the *_CONCEPT_ID fields. Classification concepts are not mapped. |
| “Maps to value” and “Value mapped from” |
Mapping to a concept that represents a Value to be placed into the VALUE_AS_CONCEPT_ID fields of the MEASUREMENT and OBSERVATION tables. |
The purpose of these mapping relationships is to allow a crosswalk between equivalent concepts to harmonize how clinical events are represented in the OMOP CDM. This is a main achievement of the Standardized Vocabularies.
“Equivalent concepts” means it carries the same meaning, and, importantly, the hierarchical descendants cover the same semantic space. If an equivalent concept is not available and the concept is not Standard, it is still mapped, but to a slightly broader concept (so-called “up-hill mappings” or semantic subsumption). For example, ICD10CM W61.51 “Bitten by goose” has no equivalent in the SNOMED vocabulary, which is generally used for standard condition concepts. Instead, it is mapped to SNOMED 217716004 “Peck by bird,” losing the context of the bird being a goose. Up-hill mappings are only used if the loss of information is considered irrelevant to standard research use cases.
Some mappings connect a source concept to more than one Standard Concept. For example, ICD9CM 070.43 “Hepatitis E with hepatic coma” is mapped to both SNOMED 235867002 “Acute hepatitis E” as well as SNOMED 72836002 “Hepatic Coma.” The reason for this is that the original source concept is a pre-coordinated combination of two conditions, hepatitis and coma, meaning that a single code simultaneously encodes multiple clinical ideas rather than expressing them separately SNOMED does not have that combination, which results in two records written to the CONDITION_OCCURRENCE table for the single ICD9CM record, one with each mapped Standard Concept.
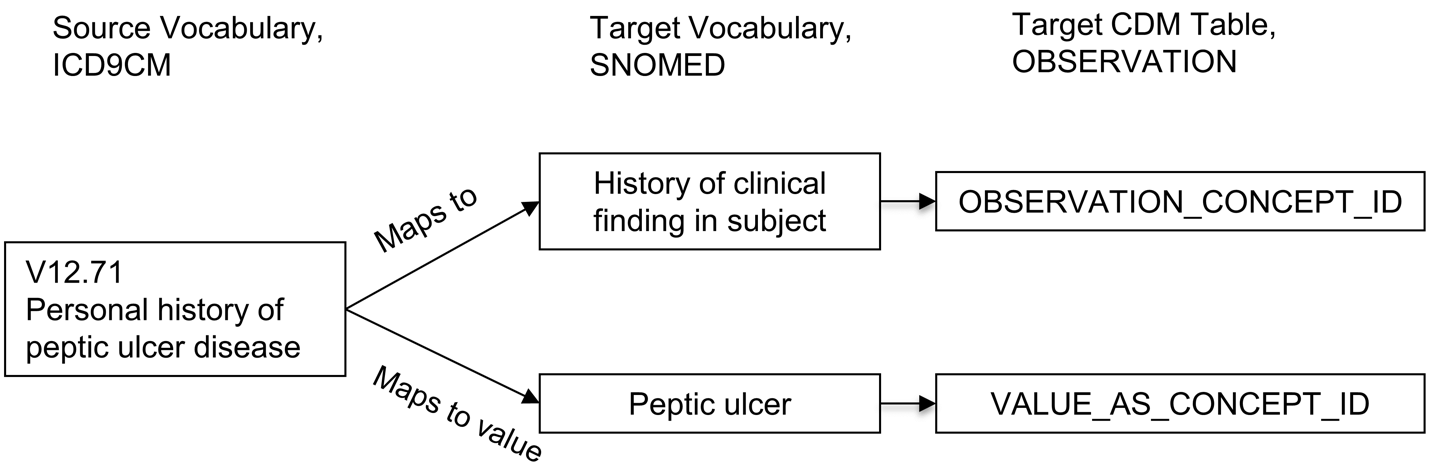
Relationships “Maps to value” have the purpose of splitting of a value for OMOP CDM tables following an entity-attribute-value (EAV) model (21). This is typically the case in the following situations:
Measurements consisting of a test and a result value
Personal or family disease history
Allergy to substance
Need for immunization
In these situations, the source concept is a combination of the attribute (test or history) and the value (test result or disease). The “Maps to” relationship maps this source to the attribute concept, and the “Maps to value” to the value concept. See Figure 5.4 for an example.
This process represents a form of controlled post-coordination within OMOP vocabularies: instead of encoding every possible combination as a new standard concept, the meaning is decomposed into two (or more) standardized elements that together fully represent the clinical event. Together, they enable more flexible, semantically rich, and extensible data modeling. By post-coordinating attribute and value concepts, OHDSI Standardized Vocabularies avoid uncontrolled growth in the number of concepts while still allowing detailed, clinically meaningful data representation and analysis. Analysts must retrieve both the CONCEPT_ID and VALUE_AS_CONCEPT_ID fields together during query building to reconstruct the complete meaning.
Mapping relationships themselves are subject to lifecycle management. Deprecated mappings (mappings with an INVALID_REASON other than NULL) are removed from active ATHENA releases but can impact longitudinal data or historical cohort definitions if not updated. Careful management of mapping versioning is crucial during vocabulary refresh cycles.
When interpreting mappings, users must be aware that not all source-to-standard mappings imply perfect semantic equivalence. Slight loss of detail, context shift, or broader aggregation may occur, particularly in uphill mappings or when representing pre-coordinated concepts. Analysts and ETL designers should validate mappings in critical analytic contexts.
5.3.13 Hierarchical Relationships and Hierarchy
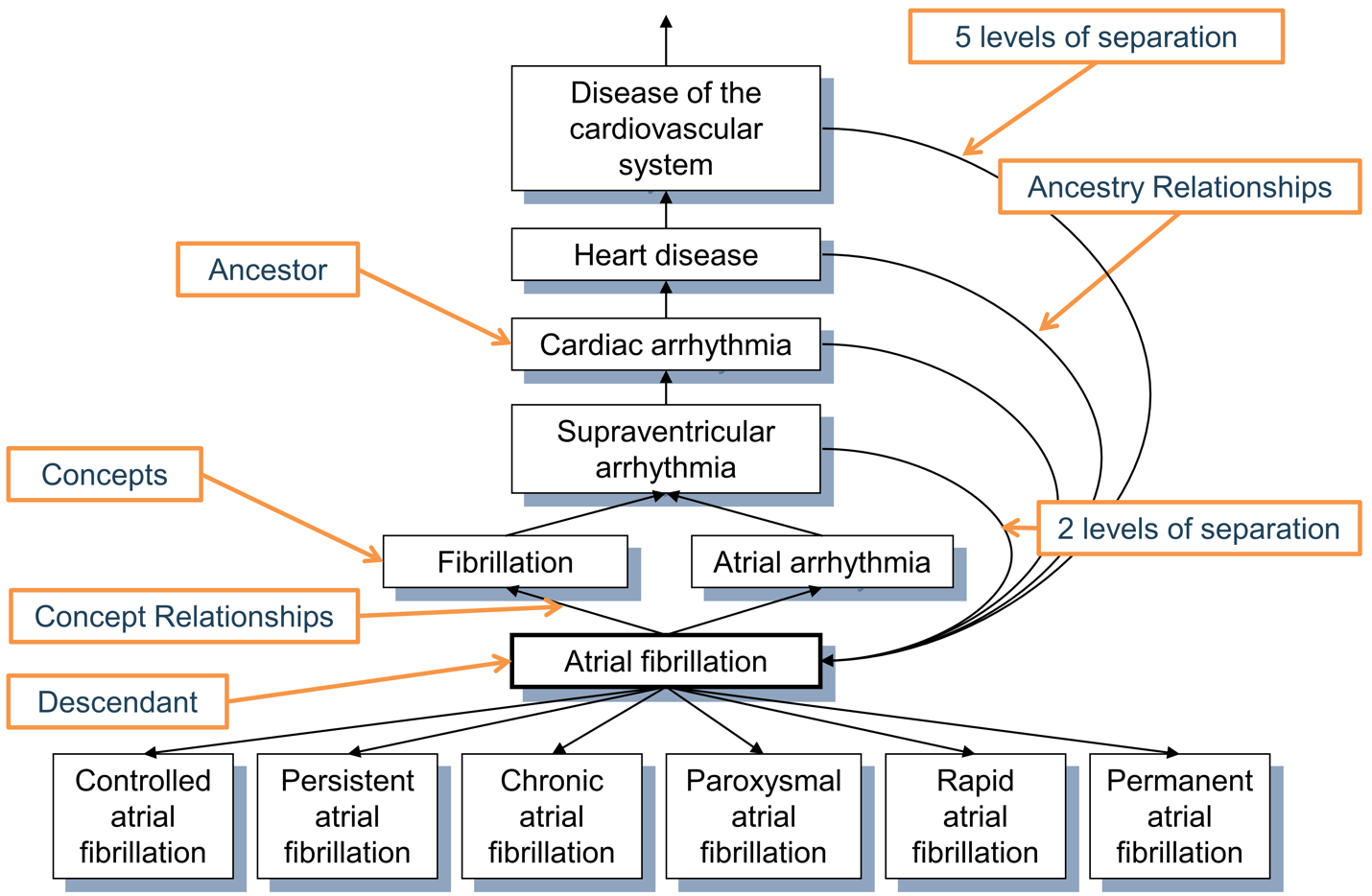
Relationships which indicate a hierarchy are defined through the “Is a” - “Subsumes” relationship pair. Hierarchical relationships are defined such that the child concept has all the attributes of the parent concept, plus one or more additional attributes or a more precisely defined attribute. For example, SNOMED 49436004 “Atrial fibrillation” is related to SNOMED 17366009 “Atrial arrhythmia” through a “Is a” relationship. Both concepts have an identical set of attributes except the type of arrhythmia, which is defined as fibrillation in one but not the other. Concepts can have more than one parent and more than one child concept. In this example, SNOMED 49436004 “Atrial fibrillation” is also an “Is a” to SNOMED 40593004 “Fibrillation.”
Within a domain, and in some cases across domains, standard and classification concepts are organized in a hierarchical structure and stored in the CONCEPT_ANCESTOR table. This allows querying and retrieving concepts and all their hierarchical descendants. These descendants have the same attributes as their ancestor, but also additional or more defined ones.
The CONCEPT_ANCESTOR table is built automatically from the CONCEPT_RELATIONSHIP table, traversing all possible concepts connected through hierarchical relationships. These are the “Is a” - “Subsumes” pairs (Figure 5.5), and other relationships connecting hierarchies across vocabularies (“SNOMED - CPT4 equivalent”, “RxNorm ingredient of”). The choice whether a relationship participates in the hierarchy constructor is defined for each relationship ID by the flag DEFINES_ANCESTRY in the RELATIONSHIP reference table. It is important to note that not all relationships with hierarchical meaning (IS_HIERARCHICAL = 1) are used for ancestry building; only those with DEFINES_ANCESTRY = 1 contribute to CONCEPT_ANCESTOR. Relationships such as “Has FDA approved indication” or “Consists of” are conceptually hierarchical but are excluded from ancestry paths to preserve clinical rigor.
CONCEPT_ANCESTOR table. Each concept is also its own descendant with both levels of separation equal to 0.
The ancestral degree, or the number of steps between ancestor and descendant, is captured in the MIN_LEVELS_OF_SEPARATION and MAX_LEVELS_OF_SEPARATION fields, defining the shortest or longest possible connection. Not all hierarchical relationships contribute equally to the levels-of-separation calculation. A step counted for the degree is determined by the IS_HIERARCHICAL flag in the RELATIONSHIP reference table for each relationship ID.
As of 2025, a high-quality comprehensive hierarchy exists only for two domains: Drug and Condition. Procedure, Measurement, and Observation domains are only partially covered and in the process of construction. The ancestry is particularly useful for the drug domain as it allows browsing all drugs with a given ingredient or members of drug classes irrespective of the country of origin, brand name or other attributes.
Users should also be aware that vocabulary updates can introduce changes to hierarchical structures, as relationships may be added, modified, or deprecated over time. Therefore, researchers are strongly encouraged to version-control their vocabulary snapshot to preserve analytic reproducibility.
5.3.14 Other Relationships
Relationships between two different vocabularies other than mapping and hierarchy relationships are typically of the type “Vocabulary A - Vocabulary B equivalent”, which is either supplied by the original source of the vocabulary or is built de-novo. They may serve as approximate mappings but often are less precise than the better curated mapping relationships. High-quality equivalence relationships (such as “Source - RxNorm equivalent”) are always duplicated by “Maps to” relationship.
Internal vocabulary relationships are usually supplied by the vocabulary provider and their quality highly depends on the vocabulary. Many of these define relationships between clinical events and can be used for information retrieval. For example, disorders of the urethra can be found by following the “Finding site of” relationship (Table 5.5):
CONCEPT_ID_1 |
CONCEPT_ID_2 |
|---|---|
| 4000504 “Urethra part” | 36713433 “Partial duplication of urethra” |
| 4000504 “Urethra part” | 433583 “Epispadias” |
| 4000504 “Urethra part” | 443533 “Epispadias, male” |
| 4000504 “Urethra part” | 4005956 “Epispadias, female” |
Internal relationships within a vocabulary may represent hierarchical (for example, “Is a”, “RxNorm ingredient of”) connections or non-hierarchical semantic associations such as anatomical location, causative agent, or associated morphology. For example, within RxNorm, relationships like “Precise ingredient of” and “Has precise ingredient” enable navigation between drug products and their precise ingredients.
5.4 Special Situations
5.4.1 Device Coding
Device concepts have no standardized coding scheme that could be used to source Standard Concepts. In many source data, devices are not even coded or contained in an external coding scheme. For this same reason, there is currently no hierarchical system available. External standards like GMDN and FDA’s UDI database have been considered but are not yet integrated. As a result, device concepts in OHDSI are mostly standard, same devices have multiple standard concepts across different vocabularies and there is no hierarchy to group terms. If you need help with devices or want to contribute talk to the OHDSI Device Workgroup and refer to {Chapter 7} of this book.
5.4.2 Coding in Oncology
Cancer data present unique modeling challenges due to the complexity of diagnoses, staging, histology, metastasis, genomic features, and treatment pathways. Please refer to the OHDSI Oncology Workgroup to learn more about conventions.
There are several mapping principles we want to cover in this chapter:
- Primary cancer diagnoses are mapped to Condition domain concepts, mostly to SNOMED CT. ICDO-3 terms are used where SNOMED coverage is insufficient.
Tumor staging, grading, and metastasis details are captured using the specialized Cancer Modifier vocabulary, which encodes structured AJCC/UICC-based elements. Mappings in Cancer Modifier are designed to ensure that cancer-related data: (1) preserve key clinical distinctions (for example, metastatic vs. localized disease), (2) support longitudinal cohort definitions (for example, new diagnosis vs. recurrence), (3) enable harmonized analytics across registries, EHRs, and claims data.
Genomic abnormalities, when available, are mapped to concepts in the OMOP Genomic vocabulary.
Oncology-specific measurements and observations, such as tumor dimensions or metastasis spread, often use post-coordination approaches - representing the entity and its result separately - to align with OMOP’s Measurement/Observation model.
Chemotherapy regimens are represented using the HemOnc vocabulary, while individual oncology drugs are mapped via RxNorm/RxNorm Extension.
More work is needed to refine mappings, remove duplicates, expand support for hematologic malignancies, and integrate molecular/genomic features.
5.4.3 Coding in Psychiatry
Psychiatric and neuropsychiatric data pose unique challenges for standardization due to the complexity of symptoms, variability of assessment tools, and evolving diagnostic frameworks. If you interested in this research talk to the OHDSI Psychiatry Workgroup.
In the OMOP model, psychiatric assessments are primarily captured within the Measurement and Observation domains, depending on whether the recorded information reflects a quantitative value or a qualitative clinical finding. Workgroup works on integrating and harmonizing Neuropsychiatric Assessment Tools, which include standardized psychometric scales, questionnaires, and structured interviews, into the Vocabularies, deduplicating terms and developing a hierarchy based on SNOMED structure to connect measurements to clinical concepts. They consider using Thesaurus of Psychological Index Terms and Human Phenotype Ontology (HPO), and real-world datasets (for example, MIMIC-IV) to inform this integration.
5.4.4 Coding for GIS, Exposomes and SDOH
Environmental context, exposomes, geographic location, and social conditions are not represented well in the OHDSI Vocabularies. If you are interested in research, talk to the OHDSI GIS Workgroup. One of the outputs of group is the OMOP GIS Vocabulary Package, which (22) delivers three coordinated vocabularies: OMOP GIS for geographic units and spatial relations, OMOP Exposome for chemicals, pollutants, toxins, and their biological targets, and OMOP SDOH for structured social-determinant indicators.
To accommodate these concepts, the package adds new domain identifiers such as Geographic Feature, Environmental Feature, Socioeconomic Feature, and Behavioral Feature. Unlike the classical OMOP domains - essentially routing flags that direct ETL to a specific CDM table - these new domains act solely as semantic groupers. They organize concepts into coherent knowledge families without prescribing storage location. Events encoded with these concepts are still recorded in the appropriate CDM tables such as EXTERNAL_EXPOSURE, OBSERVATION, or MEASUREMENT following existing conventions.
5.4.5 Microbiology and Susceptibility Coding
There are no comprehensive conventions for microbiology coding in OHDSI. You should refer to Themis conventions for the up-to-date guidance. Generally, the most common scenarios involve (1) specimen collection with a single diagnostic result (for example, Gram stain), (2) multiple lab procedures on a single sample, (3) culture-negative findings, and (4) one or more organisms identified and tested against antibiotics.
OMOP CDM supports this complexity through the MEASUREMENT, OBSERVATION, and SPECIMEN tables, with event linkages (*_EVENT_ID) connecting susceptibility results to organisms and organisms to specimens. Antibiotic susceptibility results are typically stored as LOINC-coded MEASUREMENTs with quantitative values (for example, MIC) and qualitative interpretations (for example, sensitive). When coding microbiology data you should use standard concepts from Measurement domain to populate MEASUREMENT_CONCEPT_ID (such as susceptibility test) and Meas Value domain to populate VALUE_AS_CONCEPT_ID (such as detected/not detected).
5.4.6 Survey Coding
There are no comprehensive conventions for survey coding in OHDSI. You should refer to Survey Workgroup for the up-to-date guidance. Broadly, surveys can be stored as Question-Answer pairs (separate concepts) or as pre-coordinated Question-Answer (one concept). Existing survey vocabularies, such as PPI and UK Biobank, are a mix of both. Surveys added to the Vocabularies generally should follow broad Vocabularies principles. For example, they should not contain negative information and flavors of null (not reported, not specified, etc.). If they have codes that already have standard counterparts in the Vocabularies, they should be mapped appropriately. If you want to add your survey instrument, please talk to the Survey Workgroup.
5.4.7 Flavors of NULL
Many vocabularies contain codes that represent some form of absence of information. For example, of the five gender concepts 8507 “Male,” 8532 “Female,” 8570 “Ambiguous,” 8551 “Unknown,” and 8521 “Other”, only the first two are Standard, and the other three are source concepts with no mapping. In the Standardized Vocabularies, there is intentionally no distinction why a piece of information is not available; it might be because of an active withdrawal of information by the patient, a missing value, a value that is not defined or standardized in some way, or the absence of a mapping record in CONCEPT_RELATIONSHIP. Any such concept is not mapped, which corresponds to a default mapping to the Standard Concept with the concept ID = 0.
As per Vocabularies’ principles we avoid adding new flavors of NULL to the Vocabularies and advise against using such concepts in research.
5.5 Summary
All events and administrative facts are represented in the OHDSI Standardized Vocabularies as concepts and concept relationships.
Most of these are adopted from existing coding schemes or vocabularies, while others are either extended (for example, RxNorm Extension, OMOP Extension) or developed de novo by OHDSI Vocabulary Team or community to cover missing areas.
All concepts are assigned a domain, which controls where the fact represented by the concept is stored in the CDM.
Concepts of equivalent meaning in different vocabularies are mapped to one of them, which is designated the Standard Concept. The others are source concepts. Standard concepts (“S”) are the only concepts used in analytical fields.
We strive for collaborative and transparent Vocabularies with most of the documentation located on OHDSI Vocabularies GitHub Wiki. You can get involved as a community contributor or vocabulary steward. You can contribute simple content through templates or more complex content though programmatic vocabulary development.
References
1. Reich C, Ostropolets A, Ryan P, Rijnbeek P, Schuemie M, Davydov A, et al. OHDSI Standardized Vocabularies-a large-scale centralized reference ontology for international data harmonization. J Am Med Inform Assoc. 2024 Feb 16;31(3):583–90.
2. Athena [Internet]. [cited 2025 May 23]. Available from: https://athena.ohdsi.org/search-terms/start
3. Release planning · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 23]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Release-planning
4. Standardized Vocabularies · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 23]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Standardized-Vocabularies
5. Community contribution · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 23]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Community-contribution
6. Dymshyts D. Evaluating the impact of different vocabulary versions on cohort definitions and CDM. In 2024 [cited 2025 May 23]. Available from: https://www.ohdsi.org/wp-content/uploads/2024/10/23-EvaluationConceptSets_Ddymshyts_2024_US-Dmitry-Dymshyts.pdf
7. Amos L, Anderson D, Brody S, Ripple A, Humphreys BL. UMLS users and uses: a current overview. Journal of the American Medical Informatics Association. 2020 Oct 1;27(10):1606–11.
8. De Groot R, Glaser S, Kogan A, Medlock S, Alloni A, Gabetta M, et al. ATC-to-RxNorm mappings – A comparison between OHDSI Standardized Vocabularies and UMLS Metathesaurus. International Journal of Medical Informatics. 2025 Mar;195:105777.
9. A High-Fidelity Combined ATC-Rxnorm Drug Hierarchy for Large-Scale Observational Research. In: Studies in Health Technology and Informatics [Internet]. IOS Press; 2024 [cited 2025 May 23]. Available from: https://ebooks.iospress.nl/doi/10.3233/SHTI230926
10. General Structure, Download and Use · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 23]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/General-Structure,-Download-and-Use
11. Trofymenko M, Talapova P, Williams A. OMOP GIS Vocabulary Package for Observational Studies in Health Care and Public Health. In.
12. GitHub [Internet]. [cited 2025 May 26]. Vocabulary Development Process. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Vocabulary-Development-Process
13. Quality Assurance and Control · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 26]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Quality-assurance-and-control
14. Introduction · OHDSI/Vocabulary-v5.0 Wiki · GitHub [Internet]. [cited 2025 May 26]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Introduction
15. Ostropolets A. OHDSI Vocabularies landscape assessment [Internet]. 2023. Available from: https://ohdsiorg.sharepoint.com/:w:/s/Workgroup-CommonDataModel/EQZxds1n62JIsywmDCknwtABnSb42q7hM5PwiyblXV9zDw?e=cyvxi0
16. Releases · OHDSI/Vocabulary-v5.0 [Internet]. [cited 2025 May 23]. Available from: https://github.com/OHDSI/Vocabulary-v5.0/releases
17. Matentzoglu N, Balhoff JP, Bello SM, Bizon C, Brush M, Callahan TJ, et al. A Simple Standard for Sharing Ontological Mappings (SSSOM). Database. 2022 May 25;2022:baac035.
18. OHDSI/Tantalus [Internet]. Observational Health Data Sciences and Informatics; 2024 [cited 2025 May 28]. Available from: https://github.com/OHDSI/Tantalus
19. Park Y, Yoon J, Zhuk A, Ostropolets A, You SC. Integrating Local Vocabulary into OMOP CDM: A Step-by-Step Tutorial [Internet]. 2025 [cited 2025 May 26]. Available from: http://medrxiv.org/lookup/doi/10.1101/2025.05.07.25327200
20. GitHub [Internet]. [cited 2025 May 23]. Domains. Available from: https://github.com/OHDSI/Vocabulary-v5.0/wiki/Domains
21. Dinu V, Nadkarni P. Guidelines for the effective use of entity–attribute–value modeling for biomedical databases. International Journal of Medical Informatics. 2007 Nov;76(11–12):769–79.
22. OHDSI GIS WG [Internet]. 2025 [cited 2025 May 28]. Available from: https://ohdsi.github.io/GIS/vocabulary.html