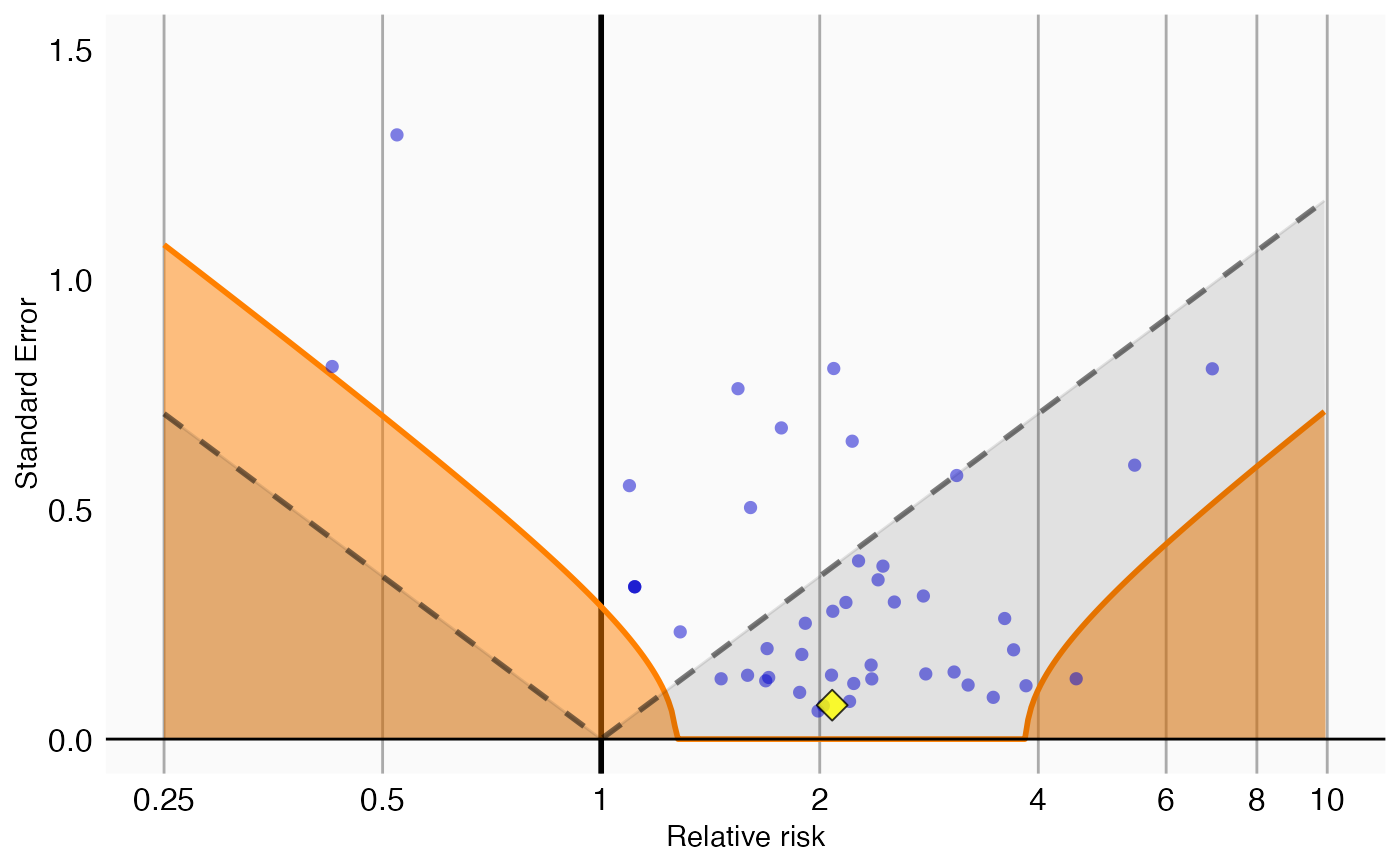
plotCalibrationEffect creates a plot showing the effect of the calibration.
Arguments
- logRrNegatives
A numeric vector of effect estimates of the negative controls on the log scale.
- seLogRrNegatives
The standard error of the log of the effect estimates of the negative controls.
- logRrPositives
Optional: A numeric vector of effect estimates of the positive controls on the log scale.
- seLogRrPositives
Optional: The standard error of the log of the effect estimates of the positive controls.
- null
An object representing the fitted null distribution as created by the
fitNullorfitMcmcNullfunctions. If not provided, a null will be fitted before plotting.- alpha
The alpha for the hypothesis test.
- xLabel
The label on the x-axis: the name of the effect estimate.
- title
Optional: the main title for the plot
- showCis
Show 95 percent credible intervals for the calibrated p = alpha boundary.
- showExpectedAbsoluteSystematicError
Show the expected absolute systematic error. If
nullis of typemcmcNullthe 95 percent credible interval will also be shown.- fileName
Name of the file where the plot should be saved, for example 'plot.png'. See the function
ggsavein the ggplot2 package for supported file formats.- xLimits
Vector of length 2 for limits of the plot x axis - defaults to 0.25, 10
- yLimits
Vector of length 2 for size limits of the y axis - defaults to 0, 1.5
Value
A Ggplot object. Use the ggsave function to save to file.
Details
Creates a plot with the effect estimate on the x-axis and the standard error on the y-axis. Negative controls are shown as blue dots, positive controls as yellow diamonds. The area below the dashed line indicated estimates with p < 0.05. The orange area indicates estimates with calibrated p < 0.05.
Examples
data(sccs)
negatives <- sccs[sccs$groundTruth == 0, ]
positive <- sccs[sccs$groundTruth == 1, ]
plotCalibrationEffect(negatives$logRr, negatives$seLogRr, positive$logRr, positive$seLogRr)
#> Warning: Removed 1 row containing missing values or values outside the scale range
#> (`geom_vline()`).