
Plot summariseMeasurementTiming results.
Source:R/plotMeasurementSummary.R
plotMeasurementSummary.RdPlot summariseMeasurementTiming results.
Usage
plotMeasurementSummary(
result,
x = "codelist_name",
y = "days_between_measurements",
plotType = "boxplot",
facet = visOmopResults::strataColumns(result),
colour = c("codelist_name"),
style = NULL
)Arguments
- result
A summarised_result object.
- x
Variable to plot on the x axis when plotType is "boxlot" or "barplot".
- y
Variable to plot, it can be "days_between_measurements" or "measurements_per_subject".
- plotType
Type of plot, either "boxplot", "barplot", or "densityplot".
- facet
Columns to facet by. See options with `visOmopResults::plotColumns(result)`. Formula input is also allowed to specify rows and columns.
- colour
Columns to color by. See options with `visOmopResults::plotColumns(result)`.
- style
Pre-defined style to apply: "default" or "darwin" - the latter just for gt and flextable. If NULL the "default" style is used.
Examples
# \donttest{
library(MeasurementDiagnostics)
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
cdm <- mockMeasurementDiagnostics()
result <- summariseMeasurementUse(
cdm = cdm,
codes = list("test_codelist" = c(3001467L, 45875977L))
)
#> → Sampling measurement table to 20000 subjects
#> → Getting measurement records based on 2 concepts.
#> → Subsetting records to the subjects and timing of interest.
#> → Getting time between records per person.
#> Summarising timings
#> ℹ The following estimates will be calculated:
#> • days_between_measurements: min, q25, median, q75, max, density
#> ! Table is collected to memory as not all requested estimates are supported on
#> the database side
#> → Start summary of data, at 2026-02-16 19:56:43.632711
#> ✔ Summary finished, at 2026-02-16 19:56:43.745347
#> → Getting measurements per subject.
#> ℹ The following estimates will be calculated:
#> • measurements_per_subject: min, q25, median, q75, max, density
#> ! Table is collected to memory as not all requested estimates are supported on
#> the database side
#> → Start summary of data, at 2026-02-16 19:56:44.363521
#> ✔ Summary finished, at 2026-02-16 19:56:44.459656
#> → Summarising results - value as number.
#> Summarising value as number
#> ℹ The following estimates will be calculated:
#> • value_as_number: min, q01, q05, q25, median, q75, q95, q99, max,
#> count_missing, percentage_missing, density
#> ! Table is collected to memory as not all requested estimates are supported on
#> the database side
#> → Start summary of data, at 2026-02-16 19:56:46.210877
#> ✔ Summary finished, at 2026-02-16 19:56:46.511755
#> → Summarising results - value as concept.
#> Summarising value as number
#> ℹ The following estimates will be calculated:
#> • value_as_concept_id: count, percentage
#> → Start summary of data, at 2026-02-16 19:56:47.246814
#> ✔ Summary finished, at 2026-02-16 19:56:47.409648
#> → Binding all diagnostic results.
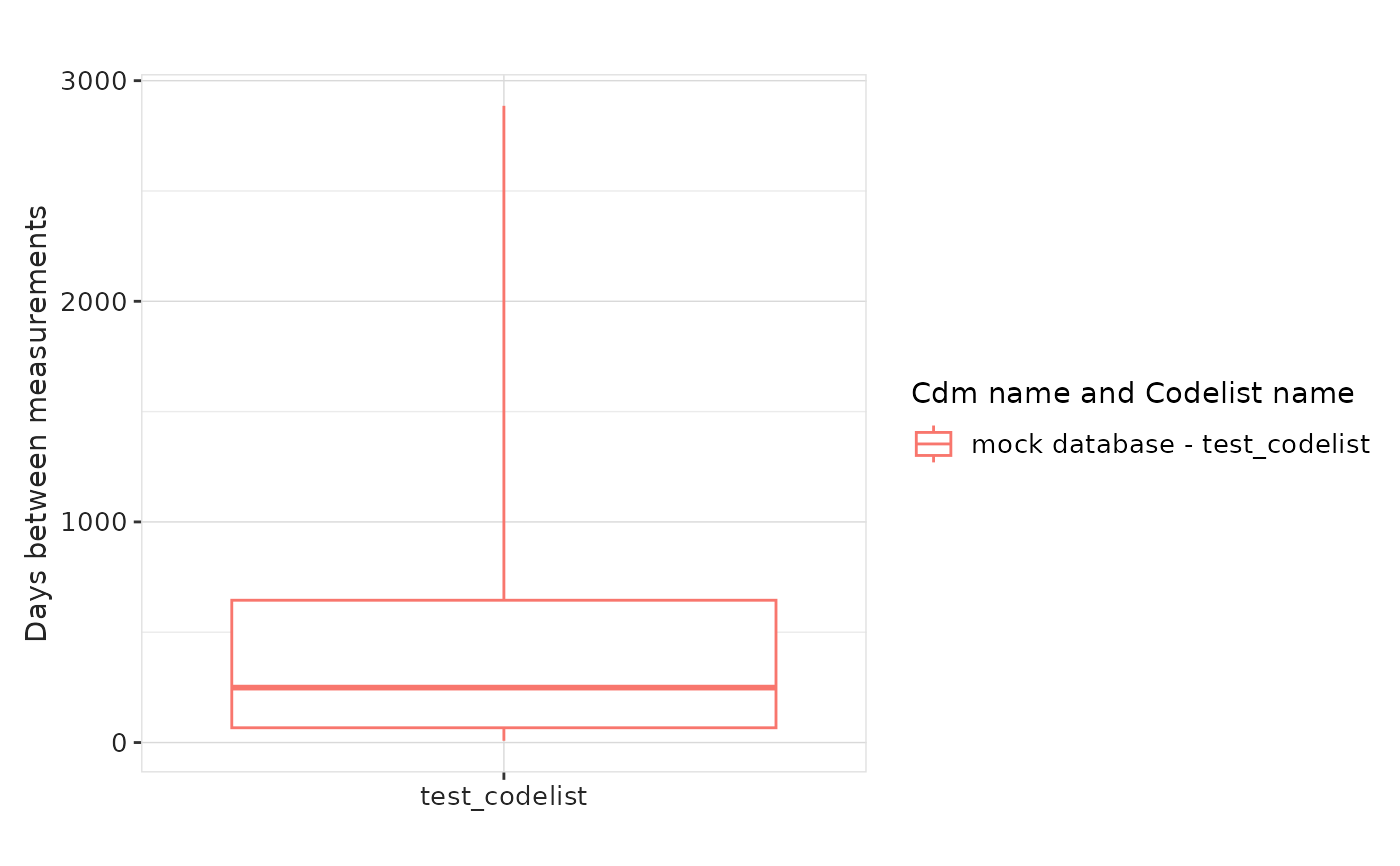
result |>
filter(variable_name == "days_between_measurements") |>
plotMeasurementSummary()
 CDMConnector::cdmDisconnect(cdm)
# }
CDMConnector::cdmDisconnect(cdm)
# }