library(duckdb)
library(dplyr)
library(omock)
library(CDMConnector)
library(omopgenerics)
library(palmerpenguins)
cdm_local <- mockCdmReference() |>
mockPerson(nPerson = 100) |>
mockObservationPeriod() |>
mockConditionOccurrence() |>
mockDrugExposure() |>
mockObservation() |>
mockMeasurement() |>
mockVisitOccurrence() |>
mockProcedureOccurrence()
con <- dbConnect(drv = duckdb())
src <- dbSource(con = con, writeSchema = "main")
cdm <- insertCdmTo(cdm = cdm_local, to = src)5 Creating a CDM reference
5.1 The OMOP CDM layout
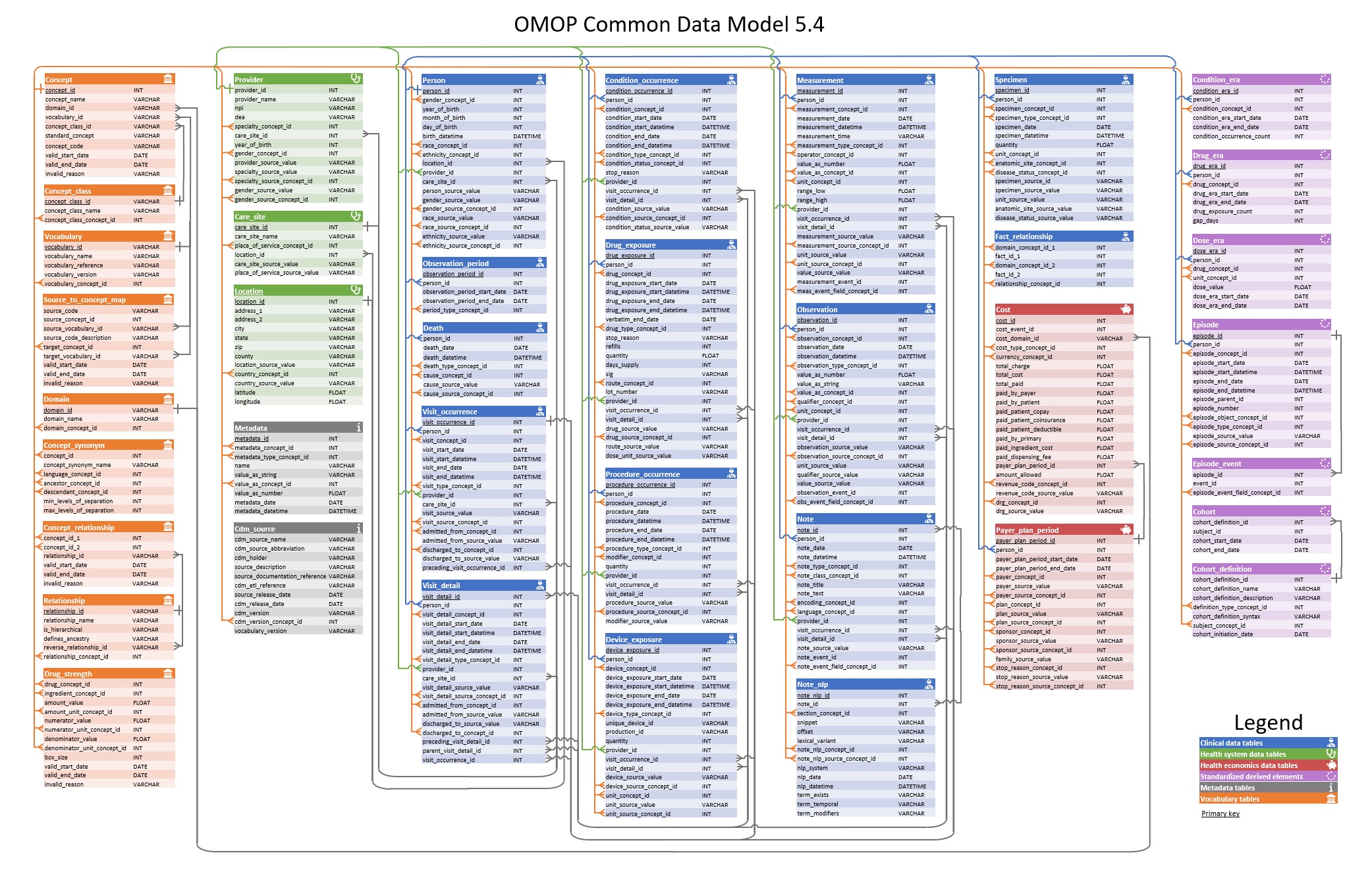
The OMOP CDM standardises the structure of healthcare data. Data is stored across a system of tables with established relationships between them. In other words, the OMOP CDM provides a relational database structure, with version 5.4 of the OMOP CDM shown below.
5.2 Creating a reference to the OMOP CDM
As we saw in Chapter 4, creating a data model in R to represent the OMOP CDM can provide a basis for analytic pipelines using the data. Luckily for us, we won’t have to create functions and methods for this ourselves. Instead, we will use the omopgenerics package which defines a data model for OMOP CDM data and the CDMConnector package which provides functions for connecting to OMOP CDM data held in a database.
To see how this works, we will use the omock package to create example data in the format of the OMOP CDM, which we will then copy to a DuckDB database created by the duckdb package.
Note that insertCdmTo() output is already a <cdm_reference> object. But how would we create this cdm reference from the connection? We can use the function cdmFromCon() from CDMConnector to create our cdm reference. Note that as well as specifying the schema containing our OMOP CDM tables, we will also specify a write schema where any database tables we create during our analysis will be stored. Often, our OMOP CDM tables will be in a schema that we only have read-access to, and we’ll have another schema where we can have write-access and where intermediate tables can be created for a given study.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cdmName = "example_data"
)cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: -• achilles tables: -• other tables: -We can also specify a write prefix and this will be used whenever permanent tables are created in the write schema. This can be useful when we’re sharing our write schema with others and want to avoid table name conflicts and easily drop tables created as part of a particular study.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "my_study_",
cdmName = "example_data"
)Note you only have to specify this writePrefix once at the connection stage, and then the cdm_reference object will store that and use it every time that you create a new table.
We can see that we now have an object that contains references to all the OMOP CDM tables. We can reference specific tables using the “$” or “[[ … ]]” operators.
cdm$person# Source: table<person> [?? x 18]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 1 8532 1971 9 1
2 2 8532 1965 10 15
3 3 8532 2000 11 4
4 4 8507 1957 1 19
5 5 8507 1980 1 25
6 6 8532 1951 4 23
7 7 8532 1967 1 17
8 8 8507 1988 4 23
9 9 8507 1953 4 16
10 10 8532 1996 5 27
# ℹ more rows
# ℹ 13 more variables: race_concept_id <int>, ethnicity_concept_id <int>,
# birth_datetime <dttm>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>cdm[["observation_period"]]# Source: table<observation_period> [?? x 5]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
person_id observation_period_s…¹ observation_period_e…² observation_period_id
<int> <date> <date> <int>
1 1 1999-06-10 2019-06-07 1
2 2 2004-04-07 2012-12-24 2
3 3 2000-11-23 2005-01-08 3
4 4 1960-09-21 1983-09-01 4
5 5 2015-05-02 2018-06-30 5
6 6 2005-11-02 2014-11-17 6
7 7 1983-01-08 2014-05-02 7
8 8 1999-05-02 2013-06-15 8
9 9 1992-08-31 2016-12-08 9
10 10 1997-12-23 2005-05-19 10
# ℹ more rows
# ℹ abbreviated names: ¹observation_period_start_date,
# ²observation_period_end_date
# ℹ 1 more variable: period_type_concept_id <int>Note that here we have first created a local version of the cdm with all the tables of interest with omock (cdm_local), then copied it to a DuckDB database, and finally created a reference to it with CDMConnector, so that we can work with the final cdm object as we normally would for one created with our own healthcare data. In that case, we would directly use cdmFromCon() with our own database information. Throughout this chapter, however, we will keep working with the mock dataset.
5.3 CDM attributes
5.3.1 CDM name
Our cdm reference will be associated with a name. By default, this name will be taken from the cdm_source_name field from the cdm_source table. We will use the function cdmName() from omopgenerics to get it.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main"
)
cdm$cdm_source |>
glimpse()Rows: ??
Columns: 10
Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
$ cdm_source_name <chr> "mock"
$ cdm_source_abbreviation <chr> NA
$ cdm_holder <chr> NA
$ source_description <chr> NA
$ source_documentation_reference <chr> NA
$ cdm_etl_reference <chr> NA
$ source_release_date <date> NA
$ cdm_release_date <date> NA
$ cdm_version <chr> "5.3"
$ vocabulary_version <chr> NAcdmName(cdm)[1] "mock"However, we can instead set this name to whatever else we want when creating our cdm reference.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cdmName = "my_cdm"
)
cdmName(cdm)[1] "my_cdm"Note that we can also get our cdm name from any of the tables in our cdm reference.
cdmName(cdm$person)[1] "my_cdm"The class of the cdm reference itself is <cdm_reference>.
class(cdm)[1] "cdm_reference"Each of the tables has class <cdm_table>. If the table is one of the standard OMOP CDM tables, it will also have class <omop_table>. This latter class is defined so that we can allow different behavior for these core tables (person, condition_occurrence, observation_period, etc.) compared to other tables that are added to the cdm reference during the course of running a study.
class(cdm$person)[1] "omop_table" "cdm_table" "tbl_duckdb_connection"
[4] "tbl_dbi" "tbl_sql" "tbl_lazy"
[7] "tbl" We can see that cdmName() is a generic function, which works for both the cdm reference as a whole and individual tables.
library(sloop)
s3_dispatch(cdmName(cdm))=> cdmName.cdm_reference
* cdmName.defaults3_dispatch(cdmName(cdm$person)) cdmName.omop_table
=> cdmName.cdm_table
cdmName.tbl_duckdb_connection
cdmName.tbl_dbi
cdmName.tbl_sql
cdmName.tbl_lazy
cdmName.tbl
* cdmName.default5.3.2 CDM version
We can also easily check the OMOP CDM version that is being used with the function cdmVersion() from omopgenerics like so:
cdmVersion(cdm)[1] "5.3"Note, the cdmVersion() function also works for <cdm_table> objects:
cdmVersion(cdm$person)[1] "5.3"Although the cdmName() and cdmVersion() functions are defined by the omopgenerics packages, these functions are re-exported in other packages and so you won’t need to load omopgenerics explicitly.
5.3.3 Source
You can get the source of a cdm object using the cdmSource() function:
cdmSource(cdm)This is a duckdb cdm sourceIn general the source is only used internally and it is an object defined by the connecting package, in this case CDMConnector and allows that all the different functions such as listSourceTables(), dropSourceTable(), readSourceTable() and compute() (that we will see later in detail) work correctly in the source backend.
In the CDMConnector case the source object contains the connection and the writing schema:
unclass(cdmSource(cdm))list()
attr(,"dbcon")<duckdb_connection ecf50 driver=<duckdb_driver dbdir=':memory:' read_only=FALSE bigint=numeric>>attr(,"write_schema")
schema
"main"
attr(,"source_type")
[1] "duckdb"For local sources for example the source object is empty:
unclass(cdmSource(cdm_local))list()
attr(,"source_type")
[1] "local"In general, the source object is not recommended to be used by the normal user, so only use it if you are proficient using the Tidy R in OMOP packages and tools.
You can easily extract the source type of your cdm as:
sourceType(cdm)[1] "duckdb"The function can also be used with a cdm_table object:
sourceType(cdm$person)[1] "duckdb"5.3.4 Retrieve the cdm reference
You can use the cdmReference() function to retrieve the cdm_reference object that a given cdm_table comes from:
cdmReference(cdm$person)── # OMOP CDM reference (duckdb) of my_cdm ─────────────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: -• achilles tables: -• other tables: -5.4 Including cohort tables in the cdm reference
A cohort is a fundamental piece in epidemiological studies. Later, we’ll see how to create cohorts in more detail in Chapter 8. For the moment, let’s just outline how we can include the reference to an existing cohort in our cdm reference. For this, we’ll use omock to add a cohort to our local cdm and upload that to a DuckDB database again.
cdm_local <- cdm_local |>
mockCohort(name = "my_study_cohort")
con <- dbConnect(drv = duckdb())
src <- dbSource(con = con, writeSchema = "main")
cdm <- insertCdmTo(cdm = cdm_local, to = src)Now we can specify we want to include this existing cohort table to our cdm object when creating our cdm reference.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cohortTables = "my_study_cohort",
cdmName = "example_data"
)
cdmcdm$my_study_cohort |>
glimpse()Rows: ??
Columns: 4
Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
$ cohort_definition_id <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ subject_id <int> 2, 3, 4, 5, 5, 5, 7, 7, 8, 8, 9, 10, 11, 12, 14, …
$ cohort_start_date <date> 2004-06-20, 2003-02-16, 1970-05-13, 2017-03-22, …
$ cohort_end_date <date> 2012-01-02, 2004-07-11, 1977-12-23, 2017-08-23, …Note that by default the cohort table won’t be included in the cdm_reference object.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cdmName = "example_data"
)
cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: -• achilles tables: -• other tables: -Even if the cohort exists in the database:
dbListTables(conn = con) [1] "cdm_source" "concept"
[3] "concept_ancestor" "concept_relationship"
[5] "concept_synonym" "condition_occurrence"
[7] "drug_exposure" "drug_strength"
[9] "measurement" "my_study_cohort"
[11] "my_study_cohort_attrition" "my_study_cohort_codelist"
[13] "my_study_cohort_set" "observation"
[15] "observation_period" "person"
[17] "procedure_occurrence" "visit_occurrence"
[19] "vocabulary" By default, only the default omop tables omopTables() will be included (if they exist) into the cdm_reference object.
5.5 Including achilles tables in the cdm reference
If we have the results tables from the Achilles package in our database, we can also include these in our cdm reference.
Just to show how this can be done, let’s upload some empty results tables in the Achilles format.
dbWriteTable(
conn = con,
name = "achilles_analysis",
value = tibble(
analysis_id = NA_integer_,
analysis_name = NA_character_,
stratum_1_name = NA_character_,
stratum_2_name = NA_character_,
stratum_3_name = NA_character_,
stratum_4_name = NA_character_,
stratum_5_name = NA_character_,
is_default = NA_character_,
category = NA_character_
)
)
dbWriteTable(
conn = con,
name = "achilles_results",
value = tibble(
analysis_id = NA_integer_,
stratum_1 = NA_character_,
stratum_2 = NA_character_,
stratum_3 = NA_character_,
stratum_4 = NA_character_,
stratum_5 = NA_character_,
count_value = NA_character_
)
)
dbWriteTable(
conn = con,
name = "achilles_results_dist",
value = tibble(
analysis_id = NA_integer_,
stratum_1 = NA_character_,
stratum_2 = NA_character_,
stratum_3 = NA_character_,
stratum_4 = NA_character_,
stratum_5 = NA_character_,
count_value = NA_character_,
min_value = NA_character_,
max_value = NA_character_,
avg_value = NA_character_,
stdev_value = NA_character_,
median_value = NA_character_,
p10_value = NA_character_,
p25_value = NA_character_,
p75_value = NA_character_,
p90_value = NA_character_
)
)We can now include these achilles tables in our cdm reference as in the previous case.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cohortTables = "my_study_cohort",
achillesSchema = "main",
cdmName = "example_data"
)cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: my_study_cohort• achilles tables: achilles_analysis, achilles_results, achilles_results_dist• other tables: -Note we specified the achillesSchema that in this case is the same as the writeSchema and cdmSchema, but each one of them can be different and point to a separate schema in our database.
5.6 Adding other tables to the cdm reference
Let’s say we have some additional local data that we want to add to our cdm reference. We can add this both to the same source (in this case a database) and to our cdm reference using insertTable() from omopgenerics (insertTable() is also re-exported in CDMConnector). We will show this with the dataset cars built-in to R.
cars |>
glimpse()Rows: 50
Columns: 2
$ speed <dbl> 4, 4, 7, 7, 8, 9, 10, 10, 10, 11, 11, 12, 12, 12, 12, 13, 13, 13…
$ dist <dbl> 2, 10, 4, 22, 16, 10, 18, 26, 34, 17, 28, 14, 20, 24, 28, 26, 34…cdm <- insertTable(cdm = cdm, name = "cars", table = cars)We can see that now this extra table has been uploaded to the database behind our cdm reference and also added to our reference.
cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: my_study_cohort• achilles tables: achilles_analysis, achilles_results, achilles_results_dist• other tables: carscdm$cars# Source: table<cars> [?? x 2]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
speed dist
<dbl> <dbl>
1 4 2
2 4 10
3 7 4
4 7 22
5 8 16
6 9 10
7 10 18
8 10 26
9 10 34
10 11 17
# ℹ more rowsIf we already had the table in the database we could have instead just assigned it to our existing cdm reference. To see this let’s upload the penguins table to our DuckDB database.
dbWriteTable(conn = con, name = "penguins", value = penguins)Once we have this table in the database, we can just read it using the readSourceTable() function.
cdm <- readSourceTable(cdm = cdm, name = "penguins")
cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: my_study_cohort• achilles tables: achilles_analysis, achilles_results, achilles_results_dist• other tables: cars, penguinsNote that omopgenerics provides these functions readSourceTable(), listSourceTables(), and dropSourceTable() for the easier management of the tables in the writeSchema.
listSourceTables(cdm = cdm) [1] "achilles_analysis" "achilles_results"
[3] "achilles_results_dist" "cars"
[5] "cdm_source" "concept"
[7] "concept_ancestor" "concept_relationship"
[9] "concept_synonym" "condition_occurrence"
[11] "drug_exposure" "drug_strength"
[13] "measurement" "my_study_cohort"
[15] "my_study_cohort_attrition" "my_study_cohort_codelist"
[17] "my_study_cohort_set" "observation"
[19] "observation_period" "penguins"
[21] "person" "procedure_occurrence"
[23] "visit_occurrence" "vocabulary" Note, not only will dropSourceTable() drop the underlying table it will also remove a reference to it in our cdm object.
cdm <- dropSourceTable(cdm = cdm, name = "penguins")
cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: my_study_cohort• achilles tables: achilles_analysis, achilles_results, achilles_results_dist• other tables: carslistSourceTables(cdm = cdm) [1] "achilles_analysis" "achilles_results"
[3] "achilles_results_dist" "cars"
[5] "cdm_source" "concept"
[7] "concept_ancestor" "concept_relationship"
[9] "concept_synonym" "condition_occurrence"
[11] "drug_exposure" "drug_strength"
[13] "measurement" "my_study_cohort"
[15] "my_study_cohort_attrition" "my_study_cohort_codelist"
[17] "my_study_cohort_set" "observation"
[19] "observation_period" "person"
[21] "procedure_occurrence" "visit_occurrence"
[23] "vocabulary" insertTable and dbWriteTable
dbWriteTable()is a function from theDBIpackage that writes a local R data frame to a database. You need to manually specify the schema and table name and it does not update the cdm reference object.insertTable()is a function from theomopgenericspackage designed for use with cdm reference objects. It writes a local table to the database and adds it to the list of tables in the cdm reference. Internally, it usesdbWriteTable()but also handles the schema and table name automatically using the writeSchema and writePrefix from the cdm reference.
In general, for studies using OMOP CDM data, you should use insertTable() rather than dbWriteTable(). It ensures the table is both written to the correct location in the database and accessible through the cdm reference object. Only use dbWriteTable() if you are confident working directly with the database and understand its structure.
Note insertTable() would also work for a local cdm reference or any other defined cdm reference source, whereas the dbWriteTable() is a database specific function.
5.7 Mutability of the cdm reference
An important characteristic of our cdm reference is that we can alter the tables in R, but the OMOP CDM data will not be affected. We will therefore only be transforming the data in our cdm object but the original datasets behind it will remain intact.
For example, let’s say we want to perform a study with only people born in 1970. For this we could filter our person table to only people born in this year.
cdm$person <- cdm$person |>
filter(year_of_birth == 1970)
cdm$person# Source: SQL [?? x 18]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 63 8532 1970 2 12
# ℹ 13 more variables: race_concept_id <int>, ethnicity_concept_id <int>,
# birth_datetime <dttm>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>From now on, when we work with our cdm reference this restriction will continue to have been applied.
cdm$person |>
tally()# Source: SQL [?? x 1]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
n
<dbl>
1 1The original OMOP CDM data itself however will remain unaffected. We can see that, indeed, if we create our reference again the underlying data is unchanged.
cdm <- cdmFromCon(
con = con,
cdmSchema = "main",
writeSchema = "main",
cdmName = "example_data"
)
cdm$person |>
tally()# Source: SQL [?? x 1]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
n
<dbl>
1 100The mutability of our cdm reference is a useful feature for studies as it means we can easily tweak our OMOP CDM data if needed. Meanwhile, leaving the underlying data unchanged is essential so that other study code can run against the data, unaffected by any of our changes.
One thing we can’t do, though, is alter the structure of OMOP CDM tables. For example, the following code would cause an error as the person table must always have the column person_id.
cdm$person <- cdm$person |>
rename("new_id" = "person_id")Error in `newOmopTable()`:
! person_id is not present in table personIn such a case we would have to call the table something else first, and then run the previous code:
cdm$person_new <- cdm$person |>
rename("new_id" = "person_id") |>
compute(name = "person_new")Now we would be allowed to have this new table as an additional table in our cdm reference, knowing it was not in the format of one of the core OMOP CDM tables.
cdm── # OMOP CDM reference (duckdb) of example_data ───────────────────────────────• omop tables: cdm_source, concept, concept_ancestor, concept_relationship,
concept_synonym, condition_occurrence, drug_exposure, drug_strength,
measurement, observation, observation_period, person, procedure_occurrence,
visit_occurrence, vocabulary• cohort tables: -• achilles tables: -• other tables: person_newThe package omopgenerics provides a comprehensive list of the required features of a valid cdm reference. You can read more about it here.
Note also that there must be a name consistency between the name of the table and the assignment in the cdm_reference object.
cdm$new_table <- cdm$person |>
compute(name = "not_new_table")Error in `[[<-`:
✖ You can't assign a table named not_new_table to new_table.
ℹ You can change the name using compute:
cdm[['new_table']] <- yourObject |>
dplyr::compute(name = 'new_table')
ℹ You can also change the name using the `name` argument in your function:
`name = 'new_table'`.5.8 Working with temporary and permanent tables
When we create new tables and our cdm reference is in a database we have a choice between using temporary or permanent tables. In most cases we can work with these interchangeably. Below we create one temporary table and one permanent table. We can see that both of these tables have been added to our cdm reference and that we can use them in the same way. Note that any new computed table will by default be temporary unless otherwise specified.
cdm$person_new_temp <- cdm$person |>
head(5) |>
compute(temporary = TRUE)cdm$person_new_permanent <- cdm$person |>
head(5) |>
compute(name = "person_new_permanent", temporary = FALSE)cdm
cdm$person_new_temp# Source: table<og_001_1762350487> [?? x 18]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 1 8532 1971 9 1
2 2 8532 1965 10 15
3 3 8532 2000 11 4
4 4 8507 1957 1 19
5 5 8507 1980 1 25
# ℹ 13 more variables: race_concept_id <int>, ethnicity_concept_id <int>,
# birth_datetime <dttm>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>cdm$person_new_permanent# Source: table<person_new_permanent> [?? x 18]
# Database: DuckDB 1.4.1 [unknown@Linux 6.11.0-1018-azure:R 4.4.1/:memory:]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 1 8532 1971 9 1
2 2 8532 1965 10 15
3 3 8532 2000 11 4
4 4 8507 1957 1 19
5 5 8507 1980 1 25
# ℹ 13 more variables: race_concept_id <int>, ethnicity_concept_id <int>,
# birth_datetime <dttm>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>One benefit of working with temporary tables is that they will be automatically dropped at the end of the session, whereas the permanent tables will be left in the database until explicitly dropped. This helps maintain the original database structure tidy and free of irrelevant data.
However, one disadvantage of using temporary tables is that we will generally accumulate more and more of them as we go (in a single R session), whereas we can overwrite permanent tables continuously. For example, if our study code contains a loop that requires a compute, we would either overwrite an intermediate permanent table 100 times or create 100 different temporary tables in the process. In the latter case we should be wary of consuming a lot of drive memory, which could lead to performance issues or even crashes.
name argument in compute()
Note that in the previous examples we explicitly specified the name of the new table and whether it must be temporary or permanent (temporary = FALSE), but we do not need to populate the temporary field explicitly as if name is left as NULL (default behavior), then the table will be temporary (temporary = TRUE), and if the name argument is populated with a character (e.g., name = “my_custom_table”), then the created table will be permanent:
cdm$person_new_temp <- cdm$person |>
compute()
cdm$person_new_permanent <- cdm$person |>
compute(name = "person_new_permanent")5.9 Disconnecting
Once we have finished our analysis we can close our connection to the database behind our cdm reference.
cdmDisconnect(cdm)5.10 Further reading
- Català M, Burn E (2025). omopgenerics: Methods and Classes for the OMOP Common Data Model. R package version 1.3.1, https://darwin-eu.github.io/omopgenerics/.
- Black A, Gorbachev A, Burn E, Català M, Nika I (2025). CDMConnector: Connect to an OMOP Common Data Model. R package version 2.2.0, https://darwin-eu.github.io/CDMConnector/.