 OHDSI GIS
WG
OHDSI GIS
WG
Gaia Toolchain
Introduction
Gaia is an integrated toolchain for combining geospatial data with OMOP Common Data Model (CDM) clinical data to enable population health research. Developed by the OHDSI GIS Working Group, Gaia provides infrastructure, software, standards, and workflows for integrating place-based datasets into patient-based health databases.
The name “Gaia” refers to the Greek personification of Earth, reflecting the toolchain’s focus on place-based health determinants and the spatial context of human health.
Mission
Improve the health of populations by generating reliable evidence from integrated geospatial and person-level health data.
Current Scope
Gaia currently supports:
- Geographic data sources: Environmental exposures, social determinants of health, demographic data, infrastructure
- Spatial data types: Polygons (census tracts, counties), points (facilities, monitoring stations), rasters (climate, pollution)
- Temporal coverage: Historical and current datasets with versioning support
- Geographic coverage: Primarily US-focused with expanding international support
- OMOP integration: External_Exposure and Location_History extension tables
- Privacy preservation: Geocoding at site, aggregated geographies, exposure values only
Limitations
Current limitations being addressed:
- Real-time data: Limited support for streaming/real-time geospatial data (planned enhancement)
- Global coverage: Majority of cataloged datasets focus on United States (expanding)
- Vocabulary coverage: Ongoing development of OMOP GIS, Exposome, and SDoH vocabularies
- Federated analytics: Multi-site federation architecture in development
- Machine learning: Limited built-in ML capabilities (planned integration)
Overview
Gaia consists of multiple interconnected components:
Core Infrastructure
gaiaDb - PostgreSQL/PostGIS database - Core data repository with all geospatial processing logic - SQL routines and PostGIS functions for spatial operations - OMOP CDM integration and exposure calculations - LinkML/JSON-LD metadata support - Transformation recipe library
gaiaCore - Multi-language connector framework - RESTful API access via PostgREST - Direct database connection support - Language-specific client libraries (R, Python) - Orchestrates functions defined in gaiaDb - Zero data processing logic (access layer only)
gaiaCatalog - Metadata catalog - Functional metadata for publicly-hosted geospatial datasets - Schema.org-compliant dataset descriptions - Automated retrieval, extraction, transformation, loading (ETL) instructions - Federated metadata sharing - Version tracking for data sources
gaiaDocker - Deployment orchestration - Coordinated image builds for entire Gaia stack - Versioned releases with docker-compose profiles - Integration with OHDSI Broadsea ecosystem - Official deployment method for all environments
Vocabulary Resources
OMOP GIS Vocabulary Package - Custom vocabularies - OMOP GIS Vocabulary (geographic entities, spatial relationships) - OMOP Exposome Vocabulary (environmental toxins, pollutants) - OMOP SDoH Vocabulary (social determinants from ADI, AHRQ, COI, EJI, SEDH, SVI, SDG, SDOHO) - Developed via Custom Vocabulary Builder (CVB) - Delta files available in TuftsCTSI/CVB
Purpose
Gaia provides a standardized, automated, reproducible, and shareable means for integrating place-based datasets into longitudinal patient health databases.
Key capabilities: - Harmonize disparate geospatial datasets into common format - Link patient locations to environmental and social exposures - Calculate spatiotemporal exposure metrics while preserving privacy - Integrate exposures into OMOP CDM for analytics - Enable federated network studies with reproducible workflows
Use Cases
Single Researcher
A researcher deploys gaiaDocker locally and gains immediate access to curated geospatial data sources:
docker compose --profile gaia up -dThey can now: - Load harmonized datasets across multiple domains (environment, demographics, SDoH) - Perform spatial joins and exploratory analyses - Create visualizations and geospatial applications - Access data via API or direct database connection
Example: Link air quality monitoring data (EPA) with social vulnerability index (CDC) to study environmental justice.
OMOP CDM Integration
A researcher with an OMOP CDM database integrates geospatial exposures:
- Geocode patient addresses locally using DeGauss (containerized, privacy-preserving)
- Spatial joins via gaiaDb SQL functions link locations to exposures
- Calculate exposures for residence periods (e.g., average PM2.5 during pregnancy)
- Populate OMOP tables: External_Exposure and Location_History
- Analytics using standard HADES tools (cohort definitions, patient-level prediction, population-level estimation)
All processing happens in gaiaDb with privacy preservation: - Only aggregated geographic identifiers (census block, ZIP) leave site - Exposure values contain no reverse-geocoding information - Standard OMOP privacy protections apply
Example: Calculate neighborhood deprivation index exposure during critical developmental windows for pediatric asthma cohort.
Federated Network Research
Gaia enables reproducible workflows across OHDSI data networks:
- Package workflow - Script entire pipeline from geocoding → exposure calculation → OMOP integration
- Containerize - Deploy via gaiaDocker with versioned releases
- Distribute - Ship to network sites with minimal configuration
- Execute - Sites run standardized workflow locally
- Aggregate - Combine summary statistics without sharing PHI
All steps include detailed provenance metadata and transformation documentation.
Example: Multi-site study examining association between greenspace exposure and mental health outcomes across diverse urban environments.
Gaia Framework
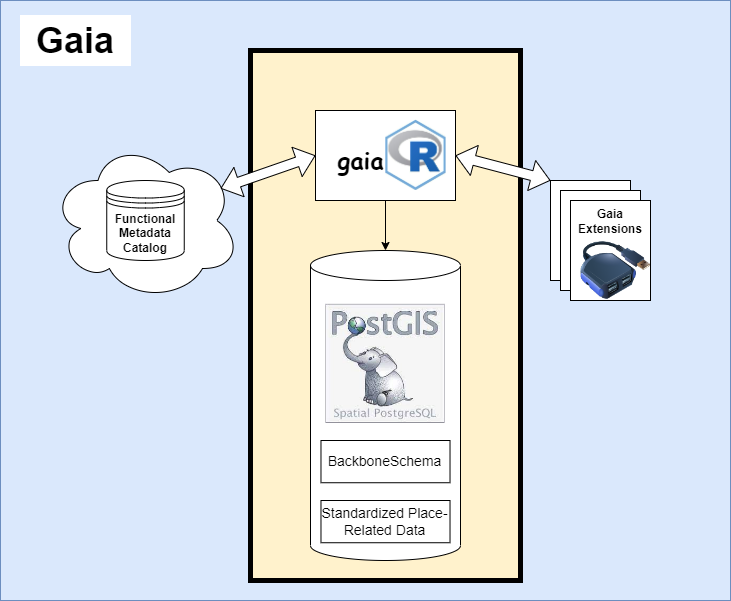
The Gaia Framework consists of three main components working together:
1. Catalog Layer
gaiaCatalog provides data discovery and metadata management:
- Descriptive metadata: Facilitates search and discovery of datasets
- Functional metadata: Machine-actionable ETL instructions
- Schema.org compliance: Standard metadata format for interoperability
- Centralized repository: Reduces duplication of ETL development effort
- Version control: Track dataset versions and transformation recipes
Data flow: External data sources → Catalog metadata → Automated ingestion
2. Core Layer
gaiaDb provides storage and processing:
- Staging database: PostgreSQL/PostGIS with LinkML-based backbone schema
- Processing engine: All SQL routines and PostGIS functions for spatial operations
- Standardized format: Entity-Attribute-Value (EAV) structure for harmonized data
- OMOP integration: Direct integration with CDM extension tables
- API exposure: PostgREST automatically generates RESTful API
gaiaCore provides multi-language access:
- Access layer: Routes requests to gaiaDb functions (zero processing logic)
- RESTful API: PostgREST-based HTTP interface
- Database connections: Direct PostgreSQL access
- Language connectors: R and Python client libraries
- OpenAPI specification: Documented API endpoints
Data flow: Raw geospatial data → gaiaDb processing → gaiaCore access → Client applications
3. Extension Layer
Specialized functionality beyond core CRUD operations:
- Visualization: Spatial data visualization tools (gaiaVis - planned)
- Analytics: Integration with HADES ecosystem
- Geocoding: DeGauss containerized geocoder integration
- Domain-specific: Custom workflows for specific research domains
Extensions interface with gaiaCore to leverage gaiaDb functionality.
Architecture Principles
Separation of Concerns
- gaiaCatalog: Discovery and metadata
- gaiaDb: Storage and processing
- gaiaCore: Multi-language access
- gaiaDocker: Deployment
Privacy by Design
- Geocoding happens at originating institution
- Only aggregated geographic identifiers shared
- Exposure values contain no location information
- OMOP privacy protections maintained
Standards Compliance
- OMOP CDM for clinical data
- Schema.org for metadata
- OGC standards for geospatial operations
- OHDSI vocabulary conventions
Reproducibility
- Version-controlled transformation recipes
- Documented data provenance
- Containerized deployments
- Open source tooling
Extensibility
- Common data model enables stable tooling
- Pluggable data sources
- Language-agnostic API access
- Modular component architecture
Getting Started
Quick Deploy
git clone https://github.com/OHDSI/gaiaDocker.git && cd gaiaDocker
docker compose --profile gaia up -dSee Get Started for detailed deployment and onboarding instructions.
Learn More
- Architecture: Technical details and data flows
- Design Principles: Schema design and functional metadata
- Vocabulary: OMOP GIS vocabulary package
- Schema Extensions: OMOP CDM extensions
Get Involved
- Join the working group
- Propose a use case
- Contribute code
- Attend Friday meetings (10 AM ET)
 OHDSI GIS
OHDSI GIS