 Gaia
Design Principles
Gaia
Design Principles
Design Rationale
Use Case Requirements
The Gaia toolchain was designed to meet specific research requirements for integrating geospatial data with patient health data:
Research Goals: - Link patient locations to environmental and social exposures over time - Harmonize disparate geospatial datasets from multiple sources - Calculate spatiotemporal exposure metrics while preserving patient privacy - Enable reproducible, federated network studies - Support diverse spatial data types (polygons, points, rasters)
Technical Requirements: - Standardized representation for heterogeneous geospatial data - Automated retrieval and ingestion of external datasets - Efficient storage and query performance for large spatial datasets - Extensible architecture supporting community growth - Integration with OMOP CDM and HADES analytics tools
Core Design Goals
The foundational goals driving Gaia’s architecture:
Extensibility
- Common data model enables stable tooling development
- Collaborative growth through shared metadata repository
- Ontology integration via OMOP vocabulary standards
- Language-agnostic access through RESTful API and database connections
Automation
- Data retrieval from external sources via functional metadata
- Standardization through transformation recipes
- Ingestion into harmonized schema
- Deployment via containerized stack (gaiaDocker)
Efficiency
- Storage design using Entity-Attribute-Value structure
- Spatial indexing for query performance
- Centralized metadata reduces duplication of ETL effort
- Modular architecture allows independent component development
Architecture Strategy
Design Approach
| Challenge | Approach |
|---|---|
| Enable extensible tooling | Implement common data model for place-related data |
| Universal representation for any geospatial data | Represent data as geometries and attributes (of geometries) |
| Efficiency with large standardized datasets | Split each data source into own pair of geometry and attribute tables |
| Static functionality for any new data | Indexing structures and parameterization treat disparate tables as functionally combined |
| Maintain source provenance and versioning | Data source and variable metadata with unique identifiers referenced throughout schema |
| Automate retrieval, ingestion, standardization | “Functional metadata” at data source and variable level, executed via gaiaDb SQL functions |
| Enable collaborative metadata growth | Host metadata centrally (gaiaCatalog), not actual data sources |
Data Pipeline
Gaia processes geospatial data through a standardized pipeline:
External Data Sources
↓
gaiaCatalog (Metadata Repository)
↓ [Functional metadata specifications]
gaiaDb ETL Functions (SQL/PostGIS)
↓ [Automated retrieval & transformation]
Harmonized Backbone Schema
↓ [GEOM_* and ATTR_* tables]
gaiaCore Access Layer
↓ [API or database connections]
Client Applications (R, Python, etc.)Pipeline stages:
- Discovery: gaiaCatalog contains metadata for external datasets
- Retrieval: gaiaDb functions fetch data from source URLs
- Transform: Transformation recipes (geom_spec, attr_spec) standardize data
- Load: Data inserted into GEOM_* and ATTR_* tables
- Index: GEOM_INDEX and ATTR_INDEX populated for parameterized querying
- Access: gaiaCore provides multi-language access to harmonized data
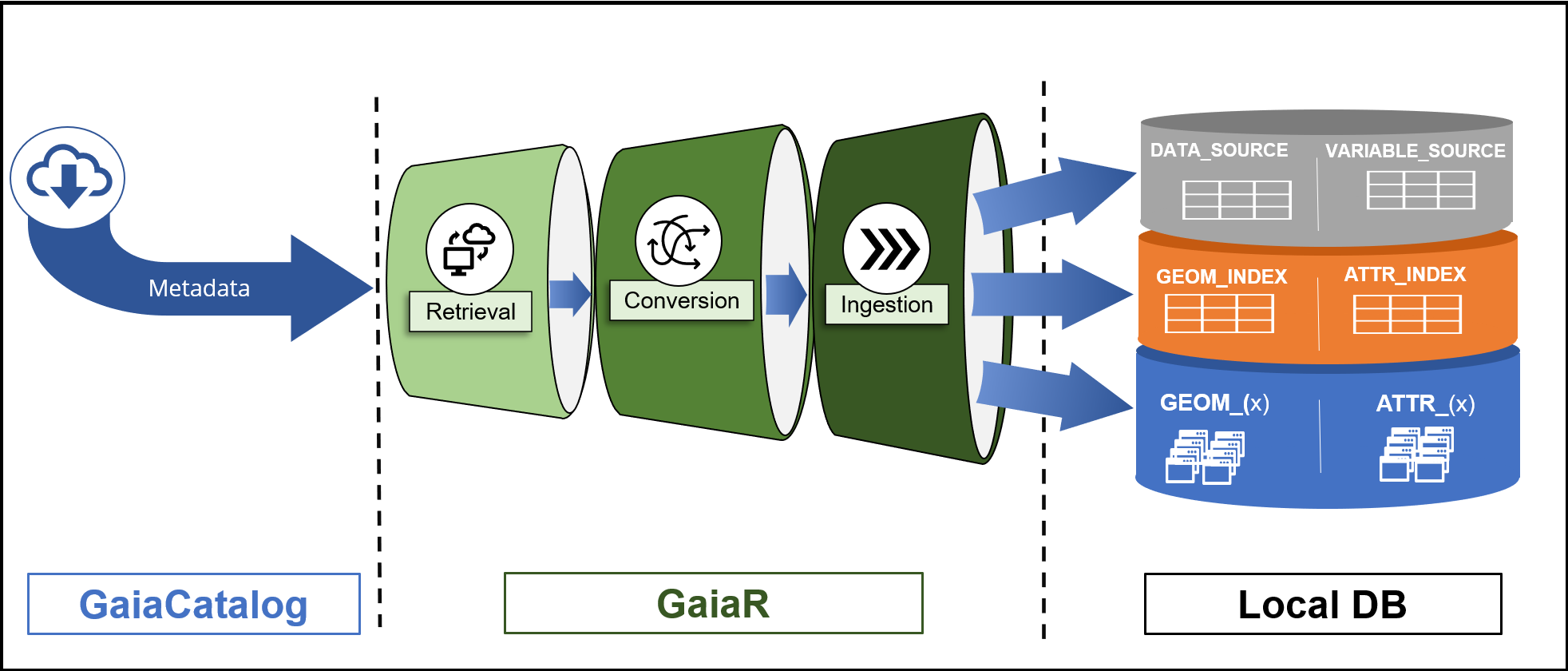
Gaia data flow from external sources through catalog metadata to harmonized storage and client access
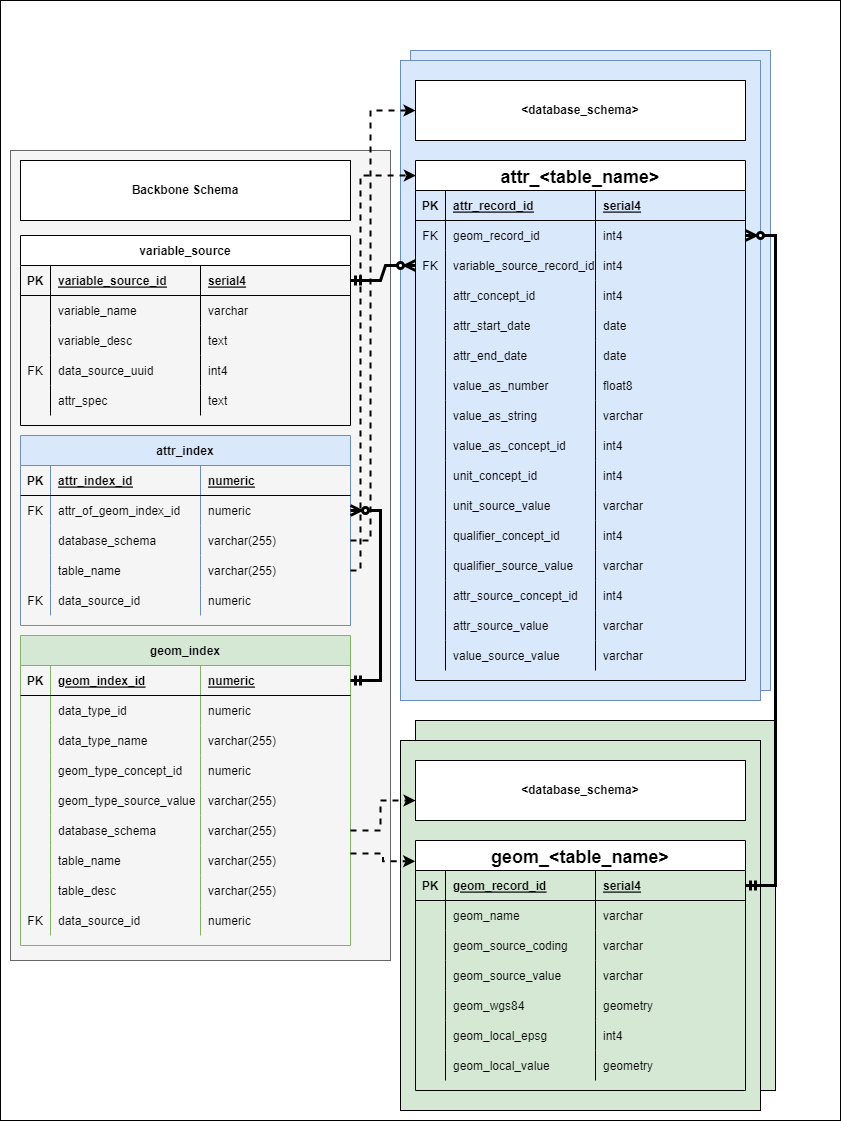
Backbone Schema
Schema Overview
The gaiaDb backbone schema consists of three distinct portions:
1. Metadata Tables
DATA_SOURCE and VARIABLE_SOURCE contain both descriptive and functional metadata:
- DATA_SOURCE: Catalogs external web-hosted datasets
- One row per unique data source
- Contains geom_spec (geometry transformation recipe)
- Links to multiple VARIABLE_SOURCE records
- VARIABLE_SOURCE: Specifies individual variables
within data sources
- Many-to-one relationship with DATA_SOURCE
- Contains attr_spec (attribute transformation recipe)
- Links to ATTR_INDEX for query mapping
2. Indexing Tables
GEOM_INDEX and ATTR_INDEX provide functional mapping to standardized data:
- Automatically populated from DATA_SOURCE metadata
- Enable parameterized querying across disparate tables
- Map data source IDs to actual table locations
- Support efficient spatial and attribute queries
Schema Diagrams
Conceptual View
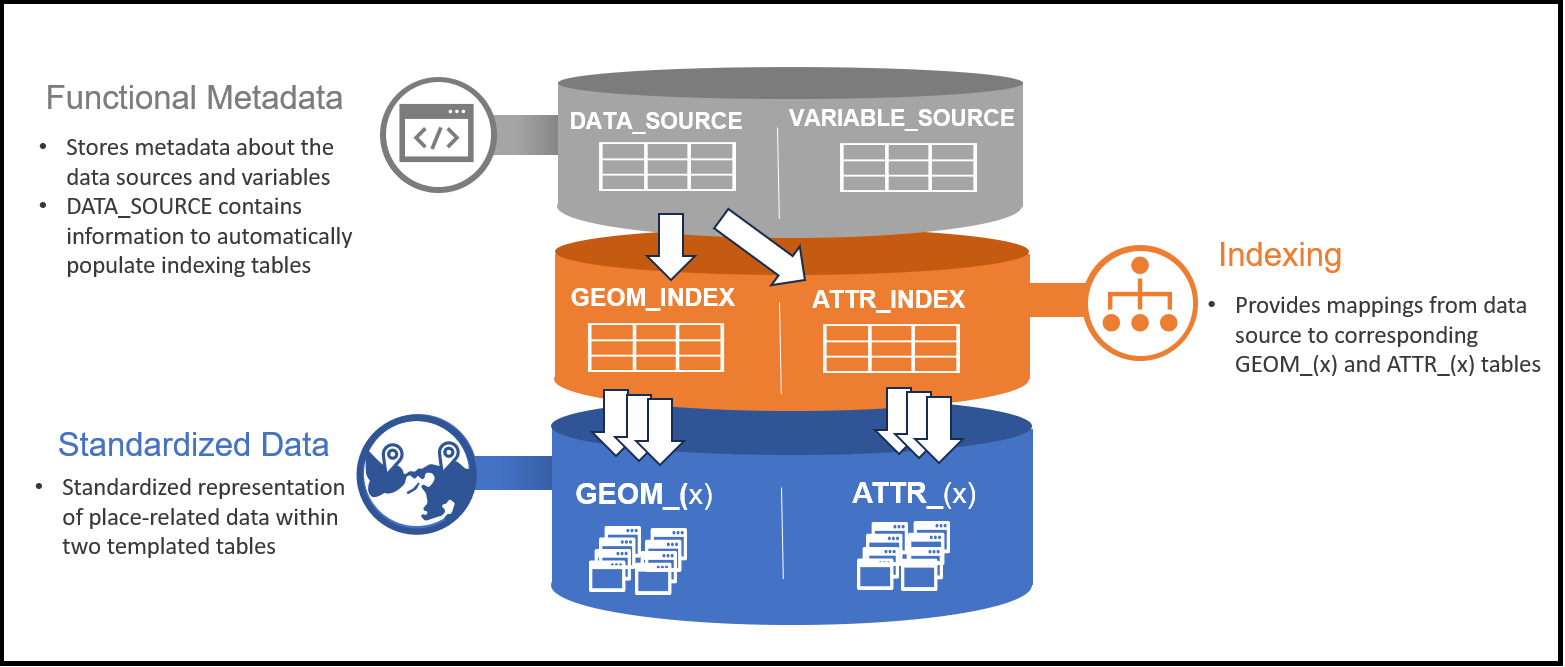
The three-tier architecture separates metadata, indexing, and data storage concerns. Metadata tables define what data exists and how to transform it. Indexing tables map logical data source identifiers to physical table locations. Standardized data tables store the harmonized geospatial data itself.
Relational Summary
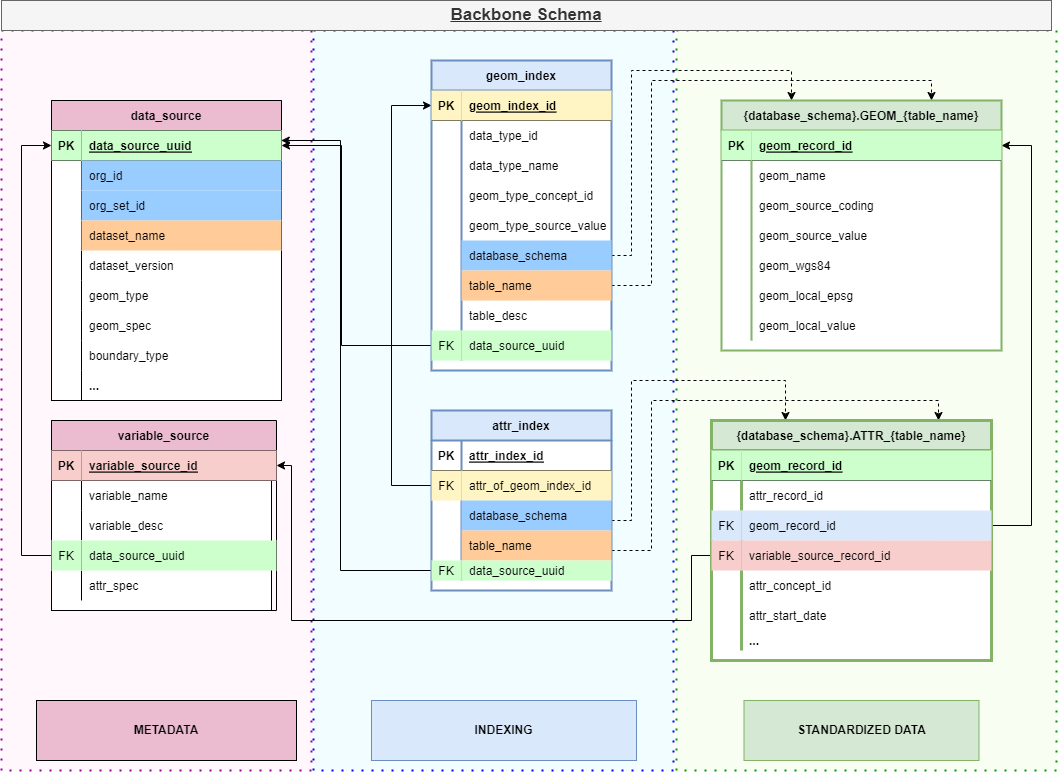
Relationships between key components: DATA_SOURCE contains functional metadata for datasets. VARIABLE_SOURCE defines variables within datasets. GEOM_INDEX and ATTR_INDEX map to physical GEOM_* and ATTR_* tables containing harmonized data.
Full Schema
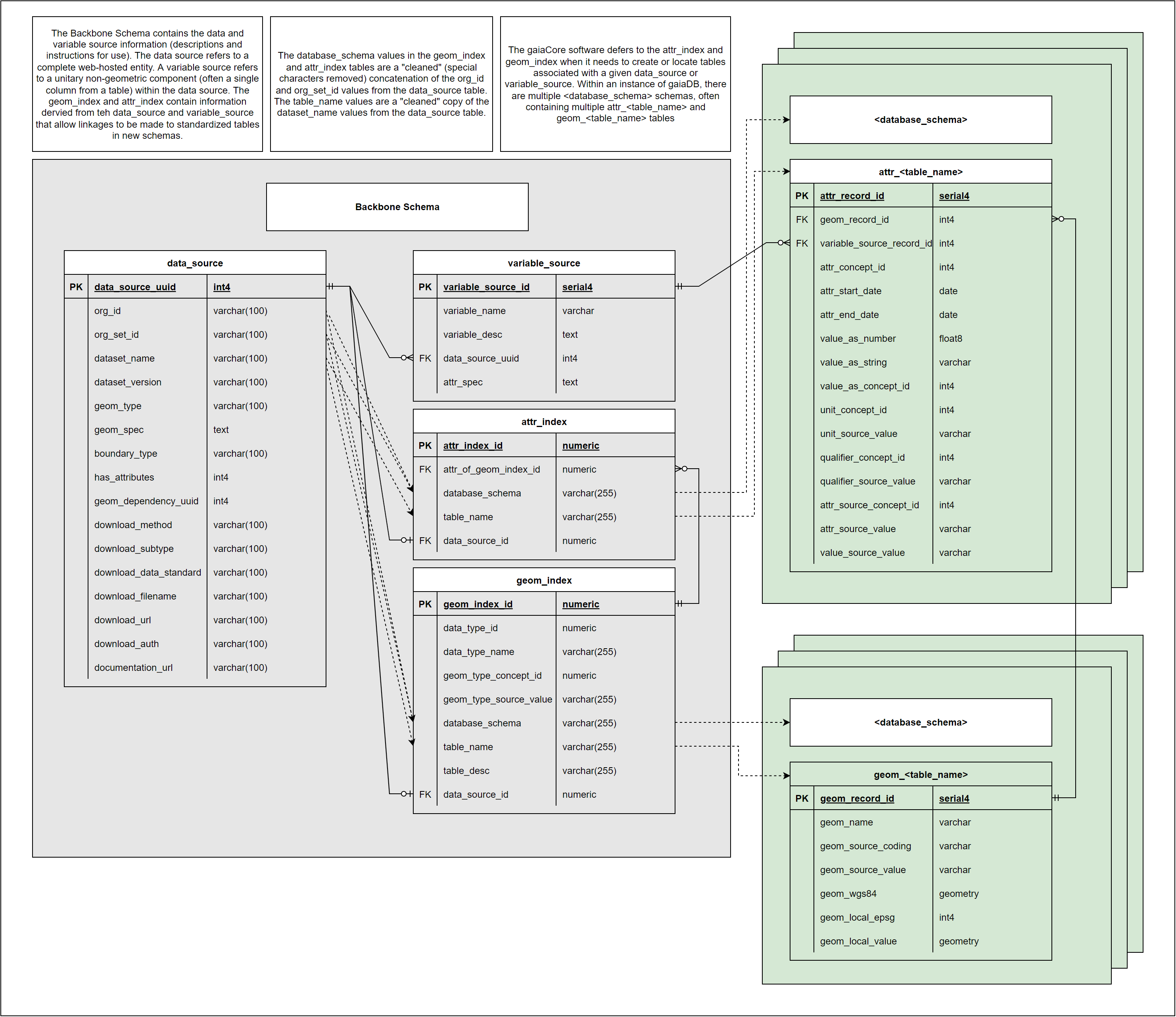
Complete entity-relationship diagram showing all tables, fields, and relationships in the gaiaDb backbone schema. Note the indexing tables that bridge metadata definitions to actual data storage.
Metadata Tables
DATA_SOURCE
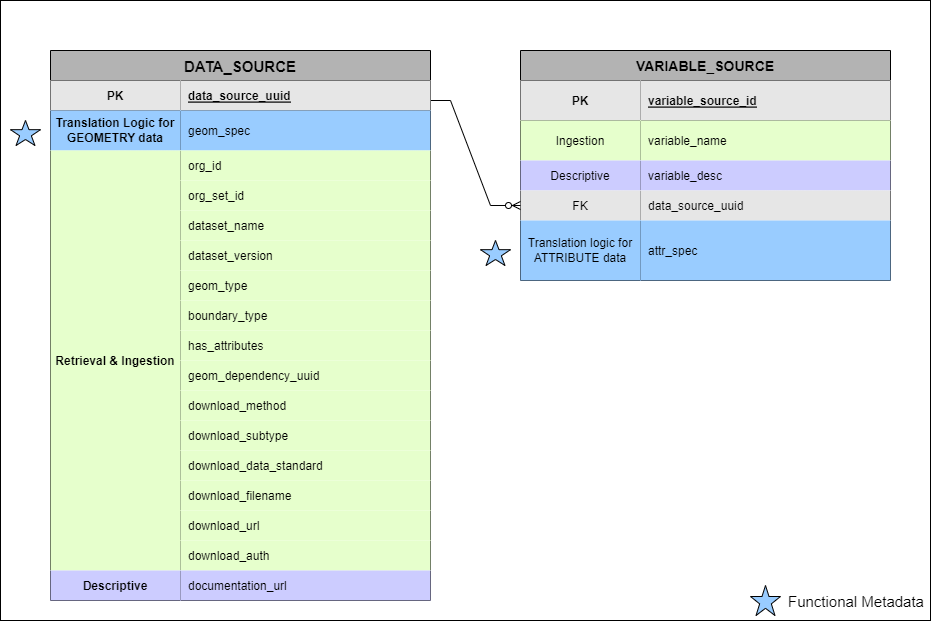
Blue stars signify functional metadata fields
Purpose: Catalog external (or local) web-hosted geospatial datasets
Key characteristics: - Every record has unique UUID - Valid when geometry type is consistent (e.g., all census tracts) - Unique combination of {Organization, Organization Set, Dataset Name} - Multiple granularities split into separate DATA_SOURCE records
Contains: - Descriptive metadata: Organization, dataset name, documentation URL, temporal coverage - Functional metadata (★): geom_spec (geometry transformation recipe), source URL, file format - Technical metadata: Schema name, table prefix, geometry type
Usage: - First step of data ingestion process - Referenced by VARIABLE_SOURCE, GEOM_INDEX, ATTR_INDEX (foreign keys) - Downloaded and ingested by gaiaDb ETL functions - Geometry translation logic stored in geom_spec field
See Backbone Specification: data_source for detailed field definitions.
VARIABLE_SOURCE
Purpose: Specify individual variables within data sources and their transformation logic
Key characteristics: - Many-to-one relationship with DATA_SOURCE - Each variable from external dataset gets own VARIABLE_SOURCE record - Links to OMOP vocabulary concepts when available
Contains: - Descriptive metadata: Variable name, description, units, temporal coverage - Functional metadata (★): attr_spec (attribute transformation recipe) - Vocabulary mapping: OMOP concept_id for standardized terminology
Usage: - Attribute translation logic stored in attr_spec field - Referenced by ATTR_INDEX for query mapping - Enables parameterized extraction of specific variables - Links geospatial attributes to OMOP vocabularies
Example: - DATA_SOURCE: “CDC Social Vulnerability Index 2020” - VARIABLE_SOURCE records: “Overall SVI Score”, “Socioeconomic Theme”, “Household Composition Theme”, etc.
See Backbone Specification: variable_source for detailed field definitions.
Indexing Tables
Indexing tables are automatically populated from DATA_SOURCE metadata and provide functional mapping to enable parameterized querying across disparate schemas and tables.

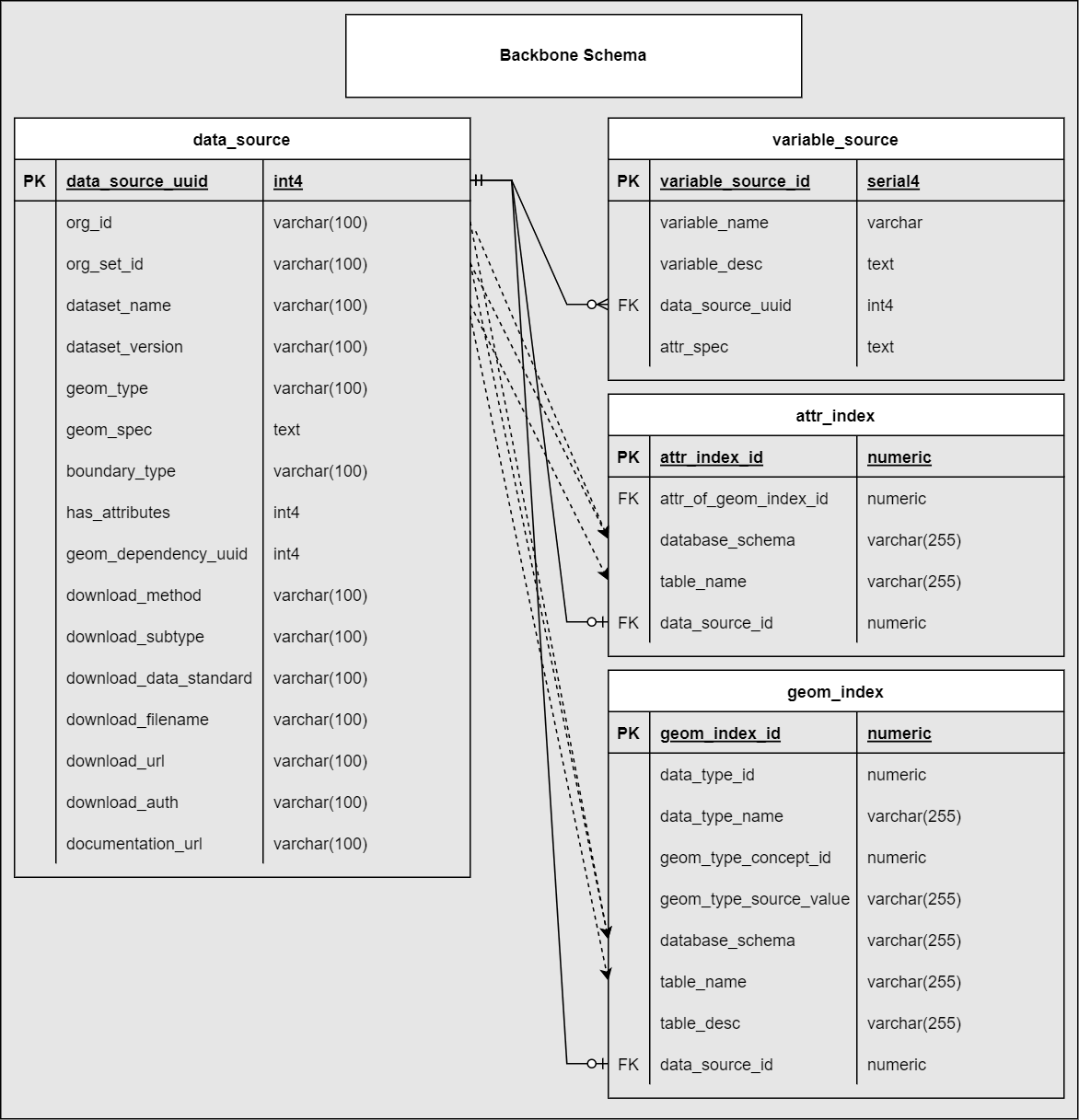
Indexing tables bridge metadata definitions to physical data storage
GEOM_INDEX
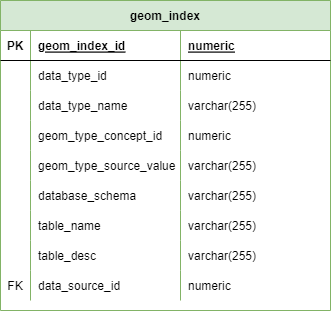
Purpose: Map data source IDs to physical geometry table locations
Key fields: - data_source_id: Links to DATA_SOURCE.uuid - schema_name: PostgreSQL schema containing geometry table - table_name: Name of GEOM_* table - geometry_type: PostGIS geometry type (POLYGON, POINT, etc.)
Usage: - Enables queries like “get all geometries for data_source_id X” - Supports parameterized spatial queries across sources - Automatically updated when new data sources ingested
See Backbone Specification: geom_index for complete definition.
ATTR_INDEX
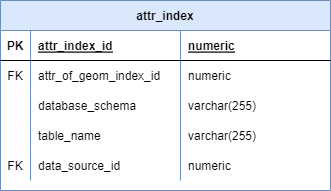
Purpose: Map variable IDs to physical attribute table locations
Key fields: - variable_source_id: Links to VARIABLE_SOURCE.uuid - data_source_id: Links to DATA_SOURCE.uuid - schema_name: PostgreSQL schema containing attribute table - table_name: Name of ATTR_* table - column_name: Specific column in ATTR table
Usage: - Enables queries like “get attribute values for variable_source_id Y” - Supports extraction of specific variables across data sources - Facilitates spatiotemporal joins for exposure calculations
See Backbone Specification: attr_index for complete definition.
Standardized Data Storage
Common Data Model Rationale
The common data model approach, proven successful for clinical data (OMOP CDM), provides:
- Interoperability: Harmonized structure enables federated research
- Extensibility: Static model supports stable tooling development
- Scalability: New data sources fit existing structure
Gaia’s approach: Represent all geospatial data as geometry objects with affiliated temporal attributes
Key transformation: Convert “wide” tables (variable columns) to “tall” Entity-Attribute-Value (EAV) structure:
Wide format:
| geom_id | county_name | pop_2020 | income_median | svi_score |
Tall format (EAV):
| geom_id | attribute_name | attribute_value | year |
| 001 | pop_2020 | 50000 | 2020 |
| 001 | income_median | 65000 | 2020 |
| 001 | svi_score | 0.45 | 2020 |Benefits: - Single standard representation for all attributes - Static column structure across all data sources - Efficient querying via indexing - Flexible addition of new variables
GEOM_TEMPLATE and ATTR_TEMPLATE
Two template tables replicate for every data source:
Each data source creates unique pair of GEOM and ATTR tables from templates
GEOM_TEMPLATE
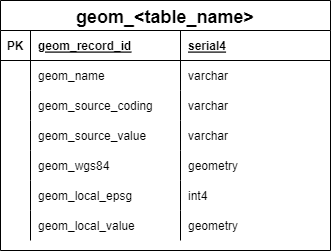
Structure: - geom_id: Unique identifier for geometry - geom: PostGIS geometry object (polygon, point, line) - geom_name: Human-readable name - parent_geom_id: For hierarchical geometries (e.g., census tract within county) - valid_from / valid_to: Temporal validity of geometry
Usage: - Template creates GEOM_* tables for each data source - Spatial indexes automatically created on geom column - Supports hierarchical spatial relationships - Temporal versioning for changing boundaries
ATTR_TEMPLATE
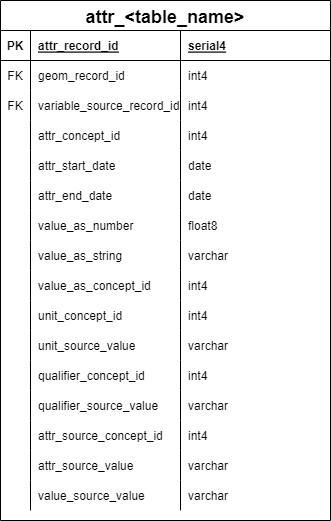
Structure: - attr_id: Unique identifier for attribute record - geom_id: Links to GEOM table - variable_source_id: Links to VARIABLE_SOURCE - attr_start_date / attr_end_date: Temporal coverage of attribute value - attr_value_as_number: Numeric attribute values - attr_value_as_string: Text attribute values - attr_value_as_concept_id: OMOP vocabulary concepts
Usage: - Template creates ATTR_* tables for each data source - EAV structure stores all attributes in standard format - Supports temporal attribute values (e.g., annual census estimates) - Links attributes to OMOP vocabularies for standardization
Functional Metadata
Functional metadata refers to machine-actionable instructions embedded in metadata records that enable automated data processing. Gaia uses functional metadata “recipes” to automate retrieval, transformation, and loading of external datasets.
Purpose
Without functional metadata: - Manual download of each dataset - Custom transformation code for each source - Undocumented, non-reproducible ETL processes - Duplication of effort across sites
With functional metadata: - Automated retrieval from source URLs - Standardized transformation via recipes - Documented, reproducible pipelines - Shared ETL logic across community
Centralized Repository
Functional metadata is hosted centrally in gaiaCatalog rather than storing actual datasets:
Benefits: - Lightweight storage (metadata only, not data) - Single source of truth for transformation recipes - Community contributions to shared catalog - Version control for metadata updates - Reduces duplication of ETL development
Workflow: 1. Dataset owner/curator creates metadata record in gaiaCatalog 2. Includes geom_spec and attr_spec transformation recipes 3. Record published to central catalog 4. Any Gaia instance can ingest dataset using metadata 5. Community improves/validates transformation recipes over time
geom_spec
Purpose: JSON specification defining how to transform source geometries into GEOM_* table structure
Contains: - File format (shapefile, GeoJSON, geopackage, etc.) - Geometry column name in source data - Geometry type expected (polygon, point, line) - Coordinate reference system (CRS) transformation - Geometry name field mapping - Hierarchical relationship mapping (if applicable) - Validity period extraction (if temporal)
Example structure (simplified):
{
"source_format": "shapefile",
"geometry_column": "geometry",
"geometry_type": "POLYGON",
"source_crs": "EPSG:4269",
"target_crs": "EPSG:4326",
"name_field": "NAME",
"parent_field": "STATEFP",
"valid_from_field": "YEAR"
}Execution: gaiaDb SQL functions parse geom_spec and execute transformation to populate GEOM_* tables.
attr_spec
Purpose: JSON specification defining how to extract and transform source attributes into ATTR_* table structure
Contains: - Source column name(s) - Data type (numeric, string, concept) - Temporal field mapping - Value transformations (e.g., unit conversions) - OMOP vocabulary mappings (if applicable) - Default values for missing data - Validation rules
Example structure (simplified):
{
"source_column": "RPL_THEMES",
"data_type": "numeric",
"temporal_field": "YEAR",
"value_transformation": "multiply_by_100",
"omop_concept_id": 2000000001,
"valid_range": [0, 1]
}Execution: gaiaDb SQL functions parse attr_spec to extract, transform, and load attributes into ATTR_* tables.
Creating Functional Metadata
Researchers can contribute new datasets by creating metadata records:
- Identify dataset: Find publicly-hosted geospatial dataset
- Create DATA_SOURCE record: Descriptive metadata + geom_spec
- Create VARIABLE_SOURCE records: One per variable + attr_spec for each
- Test transformation: Validate recipes work correctly
- Submit to catalog: Share via GitHub pull request
- Community review: Working group validates and merges
See Contributing Data Sources for detailed instructions.
Design Benefits
Reproducibility
- Version-controlled recipes: Transformation logic tracked in git
- Documented provenance: Source URLs, versions, transformation steps
- Containerized deployment: Identical environments via gaiaDocker
- Standardized vocabularies: OMOP concepts for semantic interoperability
Scalability
- Efficient storage: EAV structure with indexing
- Parallel processing: Independent data source ingestion
- Cloud-ready: Deployable on any infrastructure
- Horizontal scaling: Multiple gaiaCore instances
Community Growth
- Shared metadata: Reduces duplication of effort
- Collaborative validation: Community reviews transformation recipes
- Extensible architecture: New data sources fit existing structure
- Open source: All tools and metadata freely available
Learn More
- Gaia Overview: Introduction to Gaia toolchain
- Architecture: Technical architecture and data flows
- Vocabulary: OMOP GIS vocabulary package
- Get Started: Deploy Gaia and join the working group
 OHDSI GIS
OHDSI GIS