Using SelfControlledCohort
2022-01-06
Source:vignettes/UsingSelfControlledCohort.Rmd
UsingSelfControlledCohort.RmdThe Self Controlled Cohort method measures the association between an exposure and an outcome by comparing the number of outcomes during an unexposed time at risk to the number of outcomes during an exposed time at risk. The method is called “Self-Controlled” because each individual in the study contributes person-time to both the exposed and unexposed cohorts. Since each person contributes both exposed and unexposed time to the study only persons who experience the exposure can be used. The inputs to the analysis are the exposures and outcomes of interest, the unexposed time at risk and exposed time at risk dates for each person, and various analysis parameter settings that will be discussed in more detail.
There are seven time points to consider when defining this analysis.
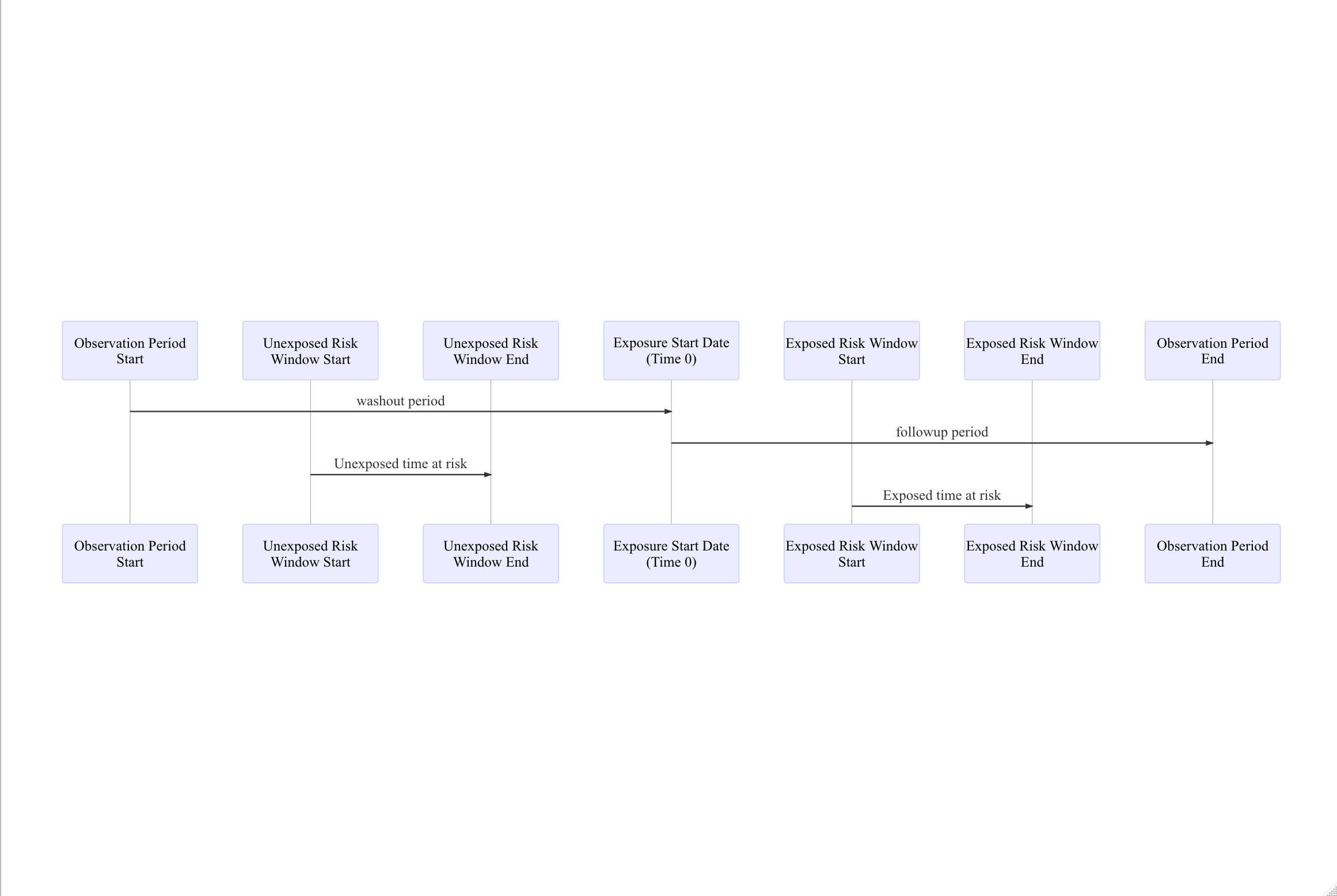
Figure: example exposure windows
For each exposure-outcome pair the following statistics are calculated:
Number of persons with the exposure
Total number of exposures (A person can experience the exposure multiple times.)
Number of outcomes during the exposed time at risk
Number of outcomes during the unexposed time at risk
Total exposed person-time at risk
Total unexposed person-time at risk
Incidence rate ratio (incidence rate during exposed time)/(incidence rate during unexposed time) -
95% confidence interval for the incidence rate ratio
The log of the incidence rate ratio - The standard error of the log incidence rate ratio
Each person’s exposure start date is taken as the index date or Time 0. The unexposed risk window is defined prior to Time 0 and represents a duration of time when the person was not exposed but could have experienced the outcome. The exposed risk window is defined as a period of time after the Time 0 and represents a duration of time when the person was exposed and could have experienced the outcome. Both of these time windows must be contained within a continuous period of observation. The details about when the risk windows start and end can be adjusted to meet the analysis specifications.
The time between the start of the observation period and Time 0 is called the washout period and a minimum allowed washout period can be set when defining the analysis. Similarly the time between Time 0 and the observation period end date is the followup period. A minimum followup period can also be set.
A common type of exposure is exposure to a drug but an exposure could be any set of characteristics that are captured in your data. We will look at a few different ways to define exposures.
Tables used in the analysis
There are two primary tables needed for the self controlled cohort analysis along with a few supporting tables that are part of the OMOP Common Data Model.
| Field | Data type |
|---|---|
| person_id | Integer |
| e xposure_concept_id | Integer |
| e xposure_start_date | Date |
| exposure_end_date | Date |
| Field | Data type |
|---|---|
| person_id | Integer |
| outcome_concept_id | Integer |
| outcome_start_date | Date |
These tables can be created manually or standard CDM tables can be used. For example by default the drug_era is used as the exposure_table and the condition_era table is used as the outcome_table. Every exposure-outcome combination in these tables will be included in the analysis unless otherwise specified. The additional CDM tables that are required for this analysis are observation_period, necessary for observation start and end dates, and person, necessary for year of birth.
A first example
The SelfControlledCohort package makes it easy to calculate the association between every drug and every condition in a CDM database. The Eunomia package provides a mini CDM that runs entirely within R that we can use for examples.
library(dplyr)
result$estimates %>%
arrange(desc(irr)) %>%
head()
#> exposureId outcomeId numPersons numExposures numOutcomesExposed
#> 1.119 705944 4048695 66 66 1
#> 1.131 705944 4134304 66 66 1
#> 1.139 705944 4230399 66 66 1
#> 1.147 705944 4296204 66 66 1
#> 1.152 705944 40479768 66 66 1
#> 1.538 740275 4146173 42 42 1
#> numOutcomesUnexposed timeAtRiskExposed timeAtRiskUnexposed irr irrLb95
#> 1.119 0 306.71869 703.4853 Inf 0.05880986
#> 1.131 0 306.71869 703.4853 Inf 0.05880986
#> 1.139 0 306.71869 703.4853 Inf 0.05880986
#> 1.147 0 306.71869 703.4853 Inf 0.05880986
#> 1.152 0 306.71869 703.4853 Inf 0.05880986
#> 1.538 0 30.78713 703.9316 Inf 0.58626854
#> irrUb95 logRr seLogRr p
#> 1.119 Inf Inf Inf NaN
#> 1.131 Inf Inf Inf NaN
#> 1.139 Inf Inf Inf NaN
#> 1.147 Inf Inf Inf NaN
#> 1.152 Inf Inf Inf NaN
#> 1.538 Inf Inf Inf NaNThe result of our analysis is a dataframe with one row per exposure-outcome pair along with all the relevant statistics. Let’s interpret the results for amoxicillin exposure (concept ID 1713332) and the outcome of Chronic sinusitis (concept ID 257012)
example <- result$estimates %>%
filter(exposureId == 1713332, outcomeId == 257012)
example %>%
tidyr::gather() %>%
mutate(value = format(round(value,1), scientific = F)) %>%
rename(column = key) %>%
knitr::kable(align = c("lr"))| column | value |
|---|---|
| exposureId | 1713332.0 |
| outcomeId | 257012.0 |
| numPersons | 2130.0 |
| numExposures | 2130.0 |
| numOutcomesExposed | 65.0 |
| numOutcomesUnexposed | 224.0 |
| timeAtRiskExposed | 277.4 |
| timeAtRiskUnexposed | 45796.0 |
| irr | 47.9 |
| irrLb95 | 35.8 |
| irrUb95 | 63.4 |
| logRr | 3.9 |
| seLogRr | 0.1 |
| p | 0.0 |
We can see that 2130 were exposed to amoxicillin. When we add up all the time before the exposure we get 4.5796044^{4} person-years. Similarly when we add up all the time after the exposure, the “Exposed time at risk”, we get 277.4428474 person-years.
The incidence of Chronic sinusitis during the “Unexposed time at risk” is
\[ \frac{224 \ \ events}{45796.0 \ \ person \ years} = 0.00489 \]
The incidence of Chronic sinusitis during the “Exposed time at risk” is
\[ \frac{65 \ \ events}{277.4 \ \ person \ years} = 0.234 \]
The incidence rate ratio is \[
\frac{234}{0.00489} = 47.9
\] with a 95% confidence interval of \([35.8, 63.4]\) See vignette("rateratio.test", package = "rateratio.test") for method details.
Custom exposure-outcome pairs
In addition to using all drugs as exposures and all conditions as outcomes we can come up with much more specific definitions of exposures and outcomes using cohorts. A cohort is a set of persons who satisfy one or more inclusion criteria for a duration of time. We can define cohorts using Atlas or by writing SQL code. If we use Atlas then the cohort will be stored in the Atlas “results” schema associated with your CDM database. If you want to write SQL then you will need write access to a schema in your CDM database.
We will simulate using cohorts created in Atlas by using the pre-built cohorts in Eunomia.
Suppose we are interested in the exposure of NSAID (cohort #4) and the outcome of GiBleed (cohort #3). Even though this is a drug-condition pair these cohorts could be defined using combinations of data elements from any domain.
result$estimates %>%
filter(exposureId == 4, outcomeId == 3) %>%
tidyr::gather() %>%
mutate(value = format(round(value,1), scientific = F)) %>%
rename(column = key) %>%
knitr::kable(align = c("lr"))| column | value |
|---|---|
| exposureId | 4.0 |
| outcomeId | 3.0 |
| numPersons | 2630.0 |
| numExposures | 2630.0 |
| numOutcomesExposed | 159.0 |
| numOutcomesUnexposed | 0.0 |
| timeAtRiskExposed | 215.7 |
| timeAtRiskUnexposed | 101570.7 |
| irr | Inf |
| irrLb95 | 20057.5 |
| irrUb95 | Inf |
| logRr | Inf |
| seLogRr | Inf |
| p | NaN |
In this case the risk of being in the GI Bleed cohort before entering the exposure cohort is zero which means our rate ratio is infinite.
Even though this example uses a drug-condition pair for the exposure and the outcome, it demonstrates how we can use any cohorts created in Atlas as exposures and outcomes in a self controlled cohort study.
Let’s demonstrate custom exposure outcome pairs using SQL by asking “Do patients tend to get more measurements in the year after a condition diagnosis than the year before?”
con <- DatabaseConnector::connect(connectionDetails)
#> Connecting using SQLite driver-- create outcome table
create table measurement_cohort as
select
person_id as subject_id,
1 as cohort_definition_id, -- treat any measurement as the same outcome
measurement_date as cohort_start_date,
measurement_date as cohort_end_date
from measurement-- create exposure table
create table condition_cohort as
select
person_id as subject_id,
2 as cohort_definition_id, -- treat any condition as the same exposure
condition_start_date as cohort_start_date,
condition_end_date as cohort_end_date
from condition_occurrenceUsing the riskWindow arguments we can set the time at risk to one year before and one year after each condition exposure.
result$estimates %>%
tidyr::gather() %>%
mutate(value = format(round(value,1), scientific = F)) %>%
rename(column = key) %>%
knitr::kable(align = c("lr"))| column | value |
|---|---|
| exposureId | 2.0 |
| outcomeId | 1.0 |
| numPersons | 2689.0 |
| numExposures | 2689.0 |
| numOutcomesExposed | 393.0 |
| numOutcomesUnexposed | 73.0 |
| timeAtRiskExposed | 6153.4 |
| timeAtRiskUnexposed | 4478.1 |
| irr | 3.9 |
| irrLb95 | 3.0 |
| irrUb95 | 5.1 |
| logRr | 1.4 |
| seLogRr | 0.1 |
| p | 0.0 |
Indeed it does appear that in Eunomia patients tend to get more tests after a diagnosis than before.
Running multiple analyses
The SelfControlledCohort package supports performing multiple analyses at once and provides functions to specify the details of each analysis to be performed.
Start by creating an sccAnalysis object for each analysis type to be performed. We will create one analysis that uses 30 day exposure windows and another with 365 day exposure windows.
sccArgs1 <- createRunSelfControlledCohortArgs(firstExposureOnly = TRUE,
firstOutcomeOnly = TRUE,
minAge = "",
maxAge = "",
studyStartDate = "",
studyEndDate = "",
addLengthOfExposureExposed = TRUE,
riskWindowStartExposed = 1,
riskWindowEndExposed = 30,
addLengthOfExposureUnexposed = TRUE,
riskWindowEndUnexposed = -1,
riskWindowStartUnexposed = -30,
hasFullTimeAtRisk = FALSE,
computeTarDistribution = TRUE,
washoutPeriod = 0,
followupPeriod = 0)
sccArgs2 <- createRunSelfControlledCohortArgs(firstExposureOnly = TRUE,
firstOutcomeOnly = TRUE,
minAge = "",
maxAge = "",
studyStartDate = "",
studyEndDate = "",
addLengthOfExposureExposed = TRUE,
riskWindowStartExposed = 1,
riskWindowEndExposed = 365,
addLengthOfExposureUnexposed = TRUE,
riskWindowEndUnexposed = -1,
riskWindowStartUnexposed = -365,
hasFullTimeAtRisk = FALSE,
computeTarDistribution = TRUE,
washoutPeriod = 0,
followupPeriod = 0)
sccAnalysis1 <- createSccAnalysis(analysisId = 1,
description = "30 day risk windows",
exposureType = NULL, # What are the valid values for this argument?
outcomeType = NULL,
runSelfControlledCohortArgs = sccArgs1)
sccAnalysis2 <- createSccAnalysis(analysisId = 2,
description = "365 day risk windows",
exposureType = NULL,
outcomeType = NULL,
runSelfControlledCohortArgs = sccArgs1)
sccAnalysisList <- list(sccAnalysis1, sccAnalysis2)Then create a list with all exposure outcome pairs to be analyzed. These can be concept IDs if you are using the condition_occurrence and drug_era tables or cohort IDs if you are using a cohort table for exposures and outcomes.
exposureOutcomeList <- list(createExposureOutcome(exposureId = 4, outcomeId = 3),
createExposureOutcome(exposureId = 1, outcomeId = 3))
The total number of rate ratios will be length(sccAnalysisList) * length(exposureOutcomesList) since every analysis will be executed for every exposure-outcome pair.
results <- runSccAnalyses(connectionDetails,
cdmDatabaseSchema = "main",
exposureTable = "cohort",
outcomeTable = "cohort",
outputFolder = "./SelfControlledCohortOutput",
sccAnalysisList = sccAnalysisList,
exposureOutcomeList = exposureOutcomeList,
analysisThreads = 1,
computeThreads = 1)
#> *** Running multiple analysis ***
summarizeAnalyses(results, "./SelfControlledCohortOutput")
#> [1] exposureId outcomeId numPersons
#> [4] numExposures numOutcomesExposed logRr
#> [7] seLogRr numOutcomesUnexposed timeAtRiskExposed
#> [10] timeAtRiskUnexposed irr irrLb95
#> [13] irrUb95 p analysisId
#> <0 rows> (or 0-length row.names)Analysis result objects are saved to the output folder and are aggregated by the summarizeAnalyses function which returns a results dataframe with one row for each result.
Correcting bias with EmpiricalCalibration
Effect estimates generated from observational data likely suffer from significant systematic bias. For example, this study design has the flaw that an exposure may look protective due to confounding by indication where patients are commonly exposed to a medication that is used to treat a comorbidity. For this reason, the SelfControlledCohort is often used with the EmpiricalCalibration package to adjust effect estimates using negative control cohorts.
See MethodEvaulation For more details on evaluating algorithms for population-level effect estimation in observational studies.