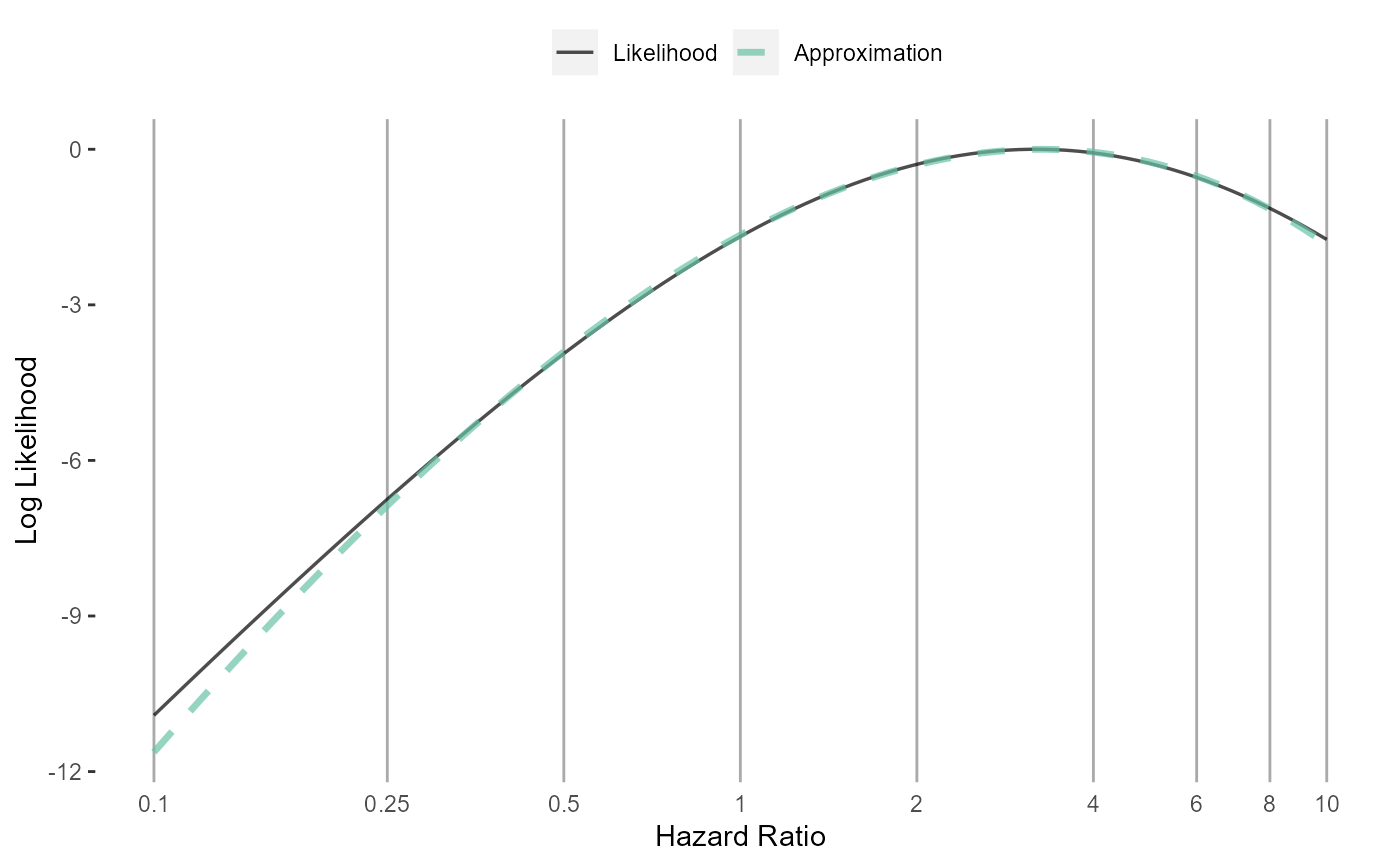
Plot the likelihood approximation
plotLikelihoodFit(
approximation,
cyclopsFit,
parameter = "x",
logScale = TRUE,
xLabel = "Hazard Ratio",
limits = c(0.1, 10),
fileName = NULL
)Arguments
- approximation
An approximation of the likelihood function as fitted using the
approximateLikelihood()function.- cyclopsFit
A model fitted using the
Cyclops::fitCyclopsModel()function.- parameter
The parameter in the
cyclopsFitobject to profile.- logScale
Show the y-axis on the log scale?
- xLabel
The title of the x-axis.
- limits
The limits on the x-axis.
- fileName
Name of the file where the plot should be saved, for example 'plot.png'. See the function ggplot2::ggsave in the ggplot2 package for supported file formats.
Value
A Ggplot object. Use the ggplot2::ggsave function to save to file.
Details
Plots the (log) likelihood and the approximation of the likelihood. Allows for reviewing the approximation.
Examples
# Simulate a single database population:
population <- simulatePopulations(createSimulationSettings(nSites = 1))[[1]]
# Approximate the likelihood:
cyclopsData <- Cyclops::createCyclopsData(Surv(time, y) ~ x + strata(stratumId),
data = population,
modelType = "cox"
)
cyclopsFit <- Cyclops::fitCyclopsModel(cyclopsData)
approximation <- approximateLikelihood(cyclopsFit,
parameter = "x",
approximation = "grid with gradients")
plotLikelihoodFit(approximation, cyclopsFit, parameter = "x")
#> Detected data following grid with gradients distribution