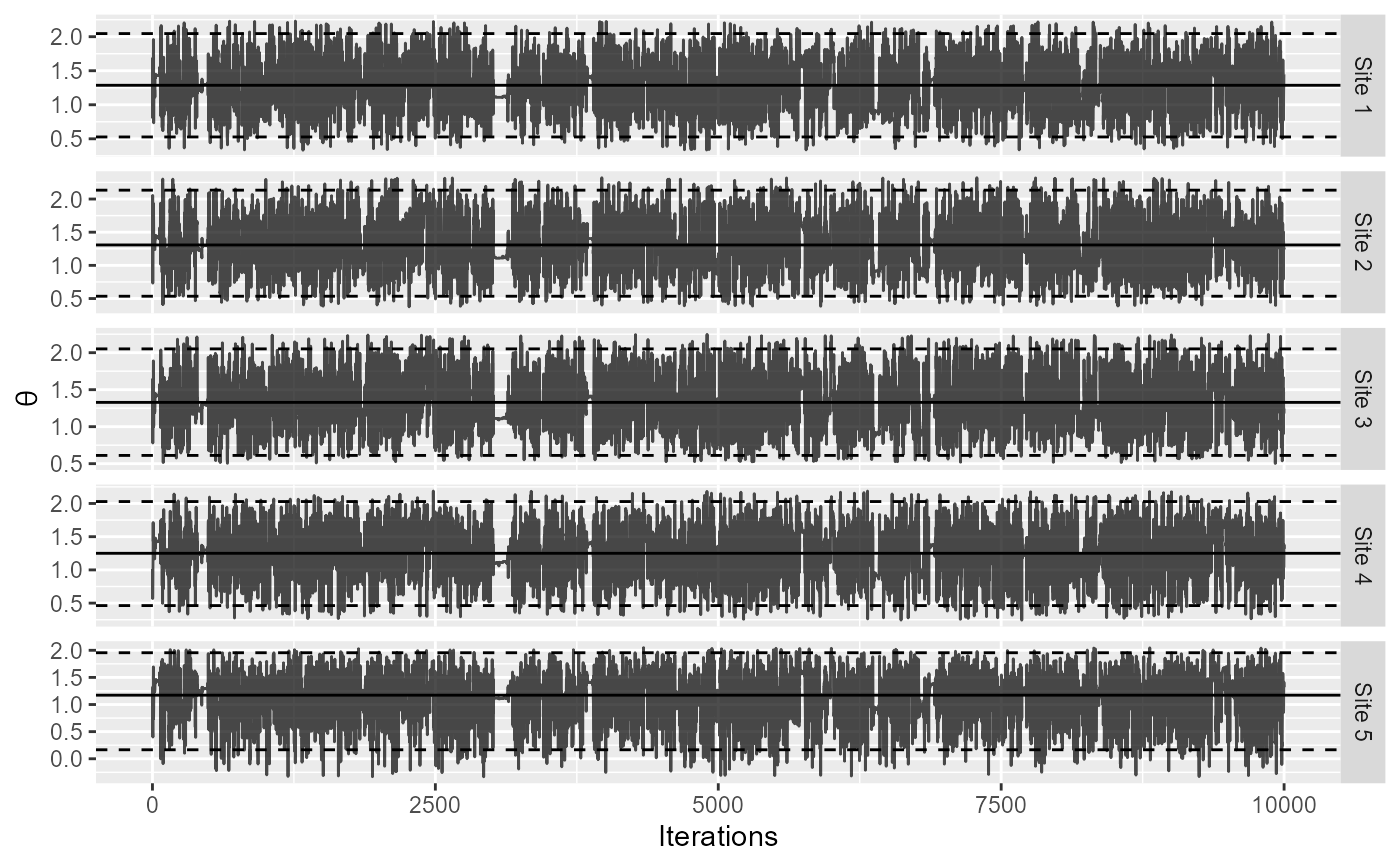
Plot MCMC trace for individual databases
plotPerDbMcmcTrace(
estimate,
showEstimate = TRUE,
dataCutoff = 0.01,
fileName = NULL
)Arguments
- estimate
An object as generated using the
computeBayesianMetaAnalysis()function.- showEstimate
Show the parameter estimates (mode) and 95 percent confidence intervals?
- dataCutoff
This fraction of the data at both tails will be removed.
- fileName
Name of the file where the plot should be saved, for example 'plot.png'. See the function ggplot2::ggsave in the ggplot2 package for supported file formats.
Value
A Ggplot object. Use the ggplot2::ggsave function to save to file.
Details
Plot the samples of the posterior distribution of the theta parameter (the estimated log hazard ratio) at each site. Samples are taken using Markov-chain Monte Carlo (MCMC).
See also
Examples
# Simulate some data for this example:
populations <- simulatePopulations()
# Fit a Cox regression at each data site, and approximate likelihood function:
fitModelInDatabase <- function(population) {
cyclopsData <- Cyclops::createCyclopsData(Surv(time, y) ~ x + strata(stratumId),
data = population,
modelType = "cox"
)
cyclopsFit <- Cyclops::fitCyclopsModel(cyclopsData)
approximation <- approximateLikelihood(cyclopsFit,
parameter = "x",
approximation = "grid with gradients")
return(approximation)
}
approximations <- lapply(populations, fitModelInDatabase)
# At study coordinating center, perform meta-analysis using per-site approximations:
estimate <- computeBayesianMetaAnalysis(approximations)
#> Detected data following grid with gradients distribution
#> Performing MCMC. This may take a while
plotPerDbMcmcTrace(estimate)