Introduction
Let’s first create a cdm reference to the Eunomia synthetic data.
library(omock)
library(CodelistGenerator)
library(PatientProfiles)
library(CohortConstructor)
library(dplyr)
library(CDMConnector)
library(duckdb)
cdm <- mockCdmFromDataset(datasetName = "GiBleed", source = "duckdb")Concept based cohort creation
A way of defining base cohorts is to identify clinical records with
codes from some pre-specified concept list. Here for example we’ll first
find codes for diclofenac and acetaminophen. We use the
getDrugIngredientCodes() function from the package
CodelistGenerator to obtain the codes for these
drugs.
drug_codes <- getDrugIngredientCodes(cdm,
name = c("acetaminophen",
"amoxicillin",
"diclofenac",
"simvastatin",
"warfarin"))
drug_codes
#>
#> - 11289_warfarin (2 codes)
#> - 161_acetaminophen (7 codes)
#> - 3355_diclofenac (1 codes)
#> - 36567_simvastatin (2 codes)
#> - 723_amoxicillin (4 codes)Now we have our codes of interest, we’ll make cohorts for each of these where cohort exit is defined as the event start date (which for these will be their drug exposure end date).
cdm$drugs <- conceptCohort(cdm,
conceptSet = drug_codes,
exit = "event_end_date",
name = "drugs")
settings(cdm$drugs)
#> # A tibble: 5 × 4
#> cohort_definition_id cohort_name cdm_version vocabulary_version
#> <int> <chr> <chr> <chr>
#> 1 1 11289_warfarin 5.3 v5.0 18-JAN-19
#> 2 2 161_acetaminophen 5.3 v5.0 18-JAN-19
#> 3 3 3355_diclofenac 5.3 v5.0 18-JAN-19
#> 4 4 36567_simvastatin 5.3 v5.0 18-JAN-19
#> 5 5 723_amoxicillin 5.3 v5.0 18-JAN-19
cohortCount(cdm$drugs)
#> # A tibble: 5 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 137 137
#> 2 2 13908 2679
#> 3 3 830 830
#> 4 4 182 182
#> 5 5 4307 2130
attrition(cdm$drugs)
#> # A tibble: 20 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 137 137 1 Initial qualif…
#> 2 1 137 137 2 Record in obse…
#> 3 1 137 137 3 Not missing re…
#> 4 1 137 137 4 Merge overlapp…
#> 5 2 14205 2679 1 Initial qualif…
#> 6 2 14205 2679 2 Record in obse…
#> 7 2 14205 2679 3 Not missing re…
#> 8 2 13908 2679 4 Merge overlapp…
#> 9 3 850 850 1 Initial qualif…
#> 10 3 830 830 2 Record in obse…
#> 11 3 830 830 3 Not missing re…
#> 12 3 830 830 4 Merge overlapp…
#> 13 4 182 182 1 Initial qualif…
#> 14 4 182 182 2 Record in obse…
#> 15 4 182 182 3 Not missing re…
#> 16 4 182 182 4 Merge overlapp…
#> 17 5 4309 2130 1 Initial qualif…
#> 18 5 4309 2130 2 Record in obse…
#> 19 5 4309 2130 3 Not missing re…
#> 20 5 4307 2130 4 Merge overlapp…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>This creates a cohort where individuals are defined by their exposure to the specified drugs, and their cohort duration is determined by the exposure end date.
Next, let’s create a cohort for individuals with bronchitis. We
define a set of codes representing bronchitis and use the
conceptCohort() function to create the cohort. Here, the
cohort exit is defined by the event start date (i.e.,
event_start_date). We set
table = "condition_occurrence" so that the records for the
provided concepts will be searched only in the
condition_occurrence table. We then set
subsetCohort = "drugs" to restrict the cohort creation to
individuals already in the drugs cohort. Additionally, we
use subsetCohortId = 1 to include only subjects from the
cohort 1 (which corresponds to individuals who have been exposed to
warfarin).
bronchitis_codes <- list(bronchitis = c(260139, 256451, 4232302))
cdm$bronchitis <- conceptCohort(cdm,
conceptSet = bronchitis_codes,
exit = "event_start_date",
name = "bronchitis",
table = "condition_occurrence",
subsetCohort = "drugs",
subsetCohortId = 1
)
cohortCount(cdm$bronchitis)
#> # A tibble: 1 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 533 130
attrition(cdm$bronchitis)
#> # A tibble: 4 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 533 130 1 Initial qualify…
#> 2 1 533 130 2 Record in obser…
#> 3 1 533 130 3 Not missing rec…
#> 4 1 533 130 4 Drop duplicate …
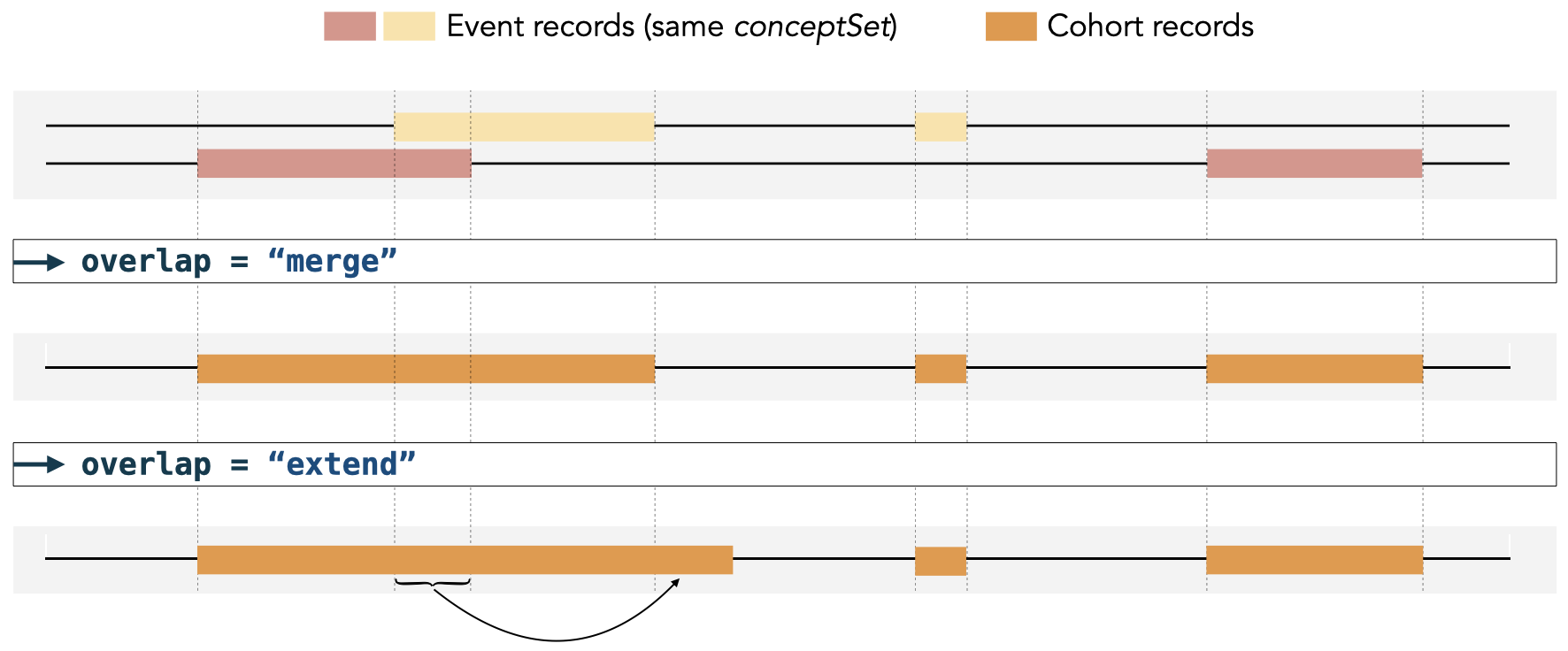
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>When some records in the cohort overlap, the cohort start date will
be set to the earliest start date. If we set
overlap = "merge", the cohort end date will be set to the
latest end date of the overlapping records.
cdm$drugs_merge <- conceptCohort(cdm,
conceptSet = drug_codes,
overlap = "merge",
name = "drugs_merge")
cdm$drugs_merge |>
attrition()
#> # A tibble: 20 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 137 137 1 Initial qualif…
#> 2 1 137 137 2 Record in obse…
#> 3 1 137 137 3 Not missing re…
#> 4 1 137 137 4 Merge overlapp…
#> 5 2 14205 2679 1 Initial qualif…
#> 6 2 14205 2679 2 Record in obse…
#> 7 2 14205 2679 3 Not missing re…
#> 8 2 13908 2679 4 Merge overlapp…
#> 9 3 850 850 1 Initial qualif…
#> 10 3 830 830 2 Record in obse…
#> 11 3 830 830 3 Not missing re…
#> 12 3 830 830 4 Merge overlapp…
#> 13 4 182 182 1 Initial qualif…
#> 14 4 182 182 2 Record in obse…
#> 15 4 182 182 3 Not missing re…
#> 16 4 182 182 4 Merge overlapp…
#> 17 5 4309 2130 1 Initial qualif…
#> 18 5 4309 2130 2 Record in obse…
#> 19 5 4309 2130 3 Not missing re…
#> 20 5 4307 2130 4 Merge overlapp…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>Alternatively, if we set overlap = "extend", the cohort
end date will be extended by summing the durations of each overlapping
record, as depicted in the image below.
cdm$drugs_extend <- conceptCohort(cdm,
conceptSet = drug_codes,
overlap = "extend",
name = "drugs_extend")
cdm$drugs_extend |>
attrition()
#> # A tibble: 30 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 137 137 1 Initial qualif…
#> 2 1 137 137 2 Record in obse…
#> 3 1 137 137 3 Not missing re…
#> 4 1 137 137 4 Add overlappin…
#> 5 1 137 137 5 Record in obse…
#> 6 1 137 137 6 Not missing re…
#> 7 2 14205 2679 1 Initial qualif…
#> 8 2 14205 2679 2 Record in obse…
#> 9 2 14205 2679 3 Not missing re…
#> 10 2 13905 2679 4 Add overlappin…
#> # ℹ 20 more rows
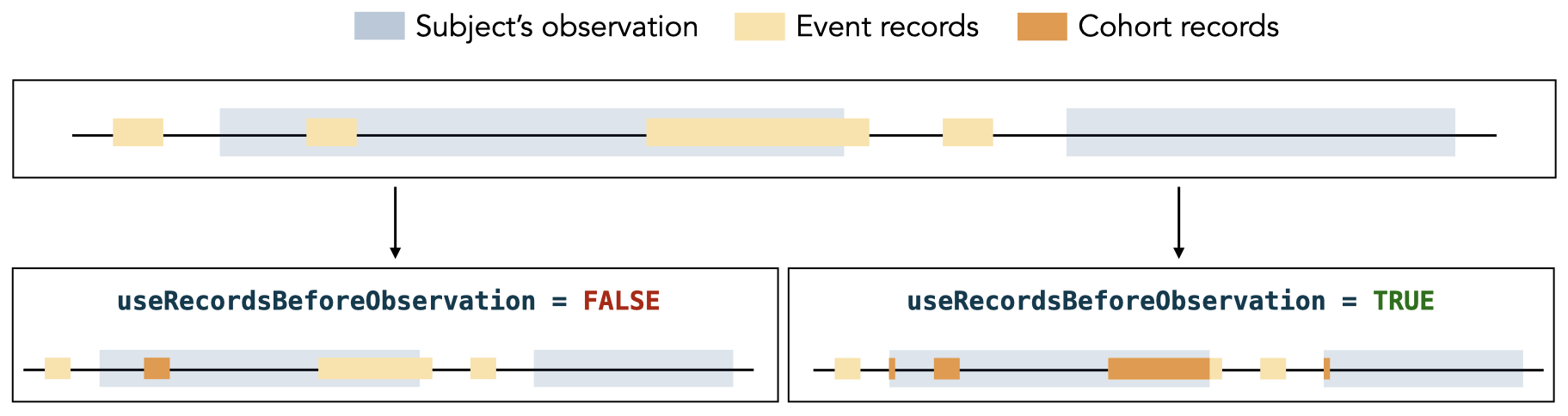
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>To create a cohort from a concept set and include records outside the
observation period, we can set
useRecordsBeforeObservation = TRUE. This option shifts
records that start before the observation period to the start of the
corresponding observation period, and trims records that start or end
after the observation period so they fall within that observation
period. This behavior is shown in the image below.
If we also want to search for the given concepts in the source
concept_id fields, rather than only the standard concept_id fields, we
can set useSourceFields = TRUE.
cdm$celecoxib <- conceptCohort(cdm,
conceptSet = list(celecoxib = 44923712),
name = "celecoxib",
useRecordsBeforeObservation = TRUE,
useSourceFields = TRUE)
cdm$celecoxib |>
glimpse()
#> Rows: ??
#> Columns: 4
#> Database: DuckDB 1.4.4 [unknown@Linux 6.14.0-1017-azure:R 4.5.2//tmp/RtmpOpDkln/file24496e097648.duckdb]
#> $ cohort_definition_id <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
#> $ subject_id <int> 578, 1733, 1929, 4403, 5152, 328, 1214, 3562, 366…
#> $ cohort_start_date <date> 2001-12-08, 2015-11-27, 1993-05-26, 1973-08-09, …
#> $ cohort_end_date <date> 2001-12-08, 2015-11-27, 1993-05-26, 1973-08-09, …Demographic based cohort creation
One base cohort we can create is based around patient demographics. Here for example we create a cohort where people enter on their 18th birthday and leave at on the day before their 66th birthday.
cdm$working_age_cohort <- demographicsCohort(cdm = cdm,
ageRange = c(18, 65),
name = "working_age_cohort")
settings(cdm$working_age_cohort)
#> # A tibble: 1 × 3
#> cohort_definition_id cohort_name age_range
#> <int> <chr> <chr>
#> 1 1 demographics 18_65
cohortCount(cdm$working_age_cohort)
#> # A tibble: 1 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 2694 2694
attrition(cdm$working_age_cohort)
#> # A tibble: 3 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 2694 2694 1 Initial qualify…
#> 2 1 2694 2694 2 Non-missing yea…
#> 3 1 2694 2694 3 Age requirement…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>We can also add an additional requirement of only people of working age with sex “female”.
cdm$female_working_age_cohort <- demographicsCohort(cdm = cdm,
ageRange = c(18, 65),
sex = "Female",
name = "female_working_age_cohort")
settings(cdm$female_working_age_cohort)
#> # A tibble: 1 × 4
#> cohort_definition_id cohort_name age_range sex
#> <int> <chr> <chr> <chr>
#> 1 1 demographics 18_65 Female
cohortCount(cdm$female_working_age_cohort)
#> # A tibble: 1 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 1373 1373
attrition(cdm$female_working_age_cohort)
#> # A tibble: 5 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 2694 2694 1 Initial qualify…
#> 2 1 2694 2694 2 Non-missing sex
#> 3 1 1373 1373 3 Sex requirement…
#> 4 1 1373 1373 4 Non-missing yea…
#> 5 1 1373 1373 5 Age requirement…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>We can also use this function to create cohorts for different combinations of age groups and sex.
cdm$age_sex_cohorts <- demographicsCohort(cdm = cdm,
ageRange = list(c(0, 17), c(18, 65), c(66,120)),
sex = c("Female", "Male"),
name = "age_sex_cohorts")
settings(cdm$age_sex_cohorts)
#> # A tibble: 6 × 4
#> cohort_definition_id cohort_name age_range sex
#> <int> <chr> <chr> <chr>
#> 1 1 demographics_1 0_17 Female
#> 2 2 demographics_2 0_17 Male
#> 3 3 demographics_3 18_65 Female
#> 4 4 demographics_4 18_65 Male
#> 5 5 demographics_5 66_120 Female
#> 6 6 demographics_6 66_120 Male
cohortCount(cdm$age_sex_cohorts)
#> # A tibble: 6 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 1373 1373
#> 2 2 1321 1321
#> 3 3 1373 1373
#> 4 4 1321 1321
#> 5 5 393 393
#> 6 6 378 378
attrition(cdm$age_sex_cohorts)
#> # A tibble: 30 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 2694 2694 1 Initial qualif…
#> 2 1 2694 2694 2 Non-missing sex
#> 3 1 1373 1373 3 Sex requiremen…
#> 4 1 1373 1373 4 Non-missing ye…
#> 5 1 1373 1373 5 Age requiremen…
#> 6 2 2694 2694 1 Initial qualif…
#> 7 2 2694 2694 2 Non-missing sex
#> 8 2 1321 1321 3 Sex requiremen…
#> 9 2 1321 1321 4 Non-missing ye…
#> 10 2 1321 1321 5 Age requiremen…
#> # ℹ 20 more rows
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>We can also specify the minimum number of days of prior observation required.
cdm$working_age_cohort_0_365 <- demographicsCohort(cdm = cdm,
ageRange = c(18, 65),
name = "working_age_cohort_0_365",
minPriorObservation = c(0,365))
settings(cdm$working_age_cohort_0_365)
#> # A tibble: 2 × 4
#> cohort_definition_id cohort_name age_range min_prior_observation
#> <int> <chr> <chr> <dbl>
#> 1 1 demographics_1 18_65 0
#> 2 2 demographics_2 18_65 365
cohortCount(cdm$working_age_cohort_0_365)
#> # A tibble: 2 × 3
#> cohort_definition_id number_records number_subjects
#> <int> <int> <int>
#> 1 1 2694 2694
#> 2 2 2694 2694
attrition(cdm$working_age_cohort_0_365)
#> # A tibble: 8 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 2694 2694 1 Initial qualify…
#> 2 1 2694 2694 2 Non-missing yea…
#> 3 1 2694 2694 3 Age requirement…
#> 4 1 2694 2694 4 Prior observati…
#> 5 2 2694 2694 1 Initial qualify…
#> 6 2 2694 2694 2 Non-missing yea…
#> 7 2 2694 2694 3 Age requirement…
#> 8 2 2694 2694 4 Prior observati…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>Measurement Cohort
Another base cohort we can create is based around patient measurements. Here for example we create a cohort of patients who have a normal BMI (BMI between 18 and 25), and a high BMI (BMI between 26 and 40). To do this you must first identify the measurement you want to look at (in this case BMI (concept id = 4245997)), the unit of measurement (kg per square-meter (concept id = 9531)), the ‘normal’ value concept (concept id = 4069590), and the concept for ‘overweight’ (concept id = 1990036). The value concept is included for the cases where the exact BMI measurement is not specified, but the BMI category (i.e. normal, overweight, obese etc) is. This means that if a record matches the value concept OR has a normal BMI score then it is included in the cohort.
cdm$cohort <- measurementCohort(
cdm = cdm,
name = "cohort",
conceptSet = list("bmi" = c(4245997)),
valueAsConcept = list("bmi_normal" = c(4069590), "bmi_overweight" = c(1990036)),
valueAsNumber = list("bmi_normal" = list("9531" = c(18, 25)), "bmi_overweight" = list("9531" = c(26, 30)))
)
attrition(cdm$cohort)
#> # A tibble: 2 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 0 0 1 Initial qualify…
#> 2 2 0 0 1 Initial qualify…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>
settings(cdm$cohort)
#> # A tibble: 2 × 4
#> cohort_definition_id cohort_name cdm_version vocabulary_version
#> <int> <chr> <chr> <chr>
#> 1 1 bmi_normal 5.3 mock
#> 2 2 bmi_overweight 5.3 mock
cdm$cohort
#> # Source: table<cohort> [?? x 4]
#> # Database: DuckDB 1.4.4 [unknown@Linux 6.14.0-1017-azure:R 4.5.2/:memory:]
#> # ℹ 4 variables: cohort_definition_id <int>, subject_id <int>,
#> # cohort_start_date <date>, cohort_end_date <date>As you can see in the above code, the concept set is the list of BMI concepts, for the “bmi_normal” cohort, the concept value is the ‘normal’ weight concept, and the values are the minimum and maximum BMI scores to be considered. Similary for the “bmi_overweight”, the concept value refers to “overweight” and the numeric range goes from minimum and maximum BMI score for that category.
It is also possible to include records outside of observation by
setting the useRecordsBeforeObservation argument to
TRUE.
cdm$cohort <- measurementCohort(
cdm = cdm,
name = "cohort",
conceptSet = list("bmi" = c(4245997)),
valueAsConcept = list("bmi_normal" = c(4069590)),
valueAsNumber = list("bmi_normal" = list("9531" = c(18, 25))),
useRecordsBeforeObservation = TRUE
)
attrition(cdm$cohort)
#> # A tibble: 1 × 7
#> cohort_definition_id number_records number_subjects reason_id reason
#> <int> <int> <int> <int> <chr>
#> 1 1 0 0 1 Initial qualify…
#> # ℹ 2 more variables: excluded_records <int>, excluded_subjects <int>
settings(cdm$cohort)
#> # A tibble: 1 × 4
#> cohort_definition_id cohort_name cdm_version vocabulary_version
#> <int> <chr> <chr> <chr>
#> 1 1 bmi_normal 5.3 mock
cdm$cohort
#> # Source: table<cohort> [?? x 4]
#> # Database: DuckDB 1.4.4 [unknown@Linux 6.14.0-1017-azure:R 4.5.2/:memory:]
#> # ℹ 4 variables: cohort_definition_id <int>, subject_id <int>,
#> # cohort_start_date <date>, cohort_end_date <date>Death cohort
Another base cohort we can make is one with individuals who have died. For this we’ll simply be finding those people in the OMOP CDM death table and creating a cohort with them.
cdm$death_cohort <- deathCohort(cdm = cdm,
name = "death_cohort")To create a cohort of individuals who have died, but restrict it to
those already part of another cohort (e.g., the “drugs” cohort), you can
use subsetCohort = "drugs". This ensures that only
individuals from the “drugs” cohort are included in the death
cohort.
cdm$death_drugs <- deathCohort(cdm = cdm,
name = "death_drugs",
subsetCohort = "drugs")