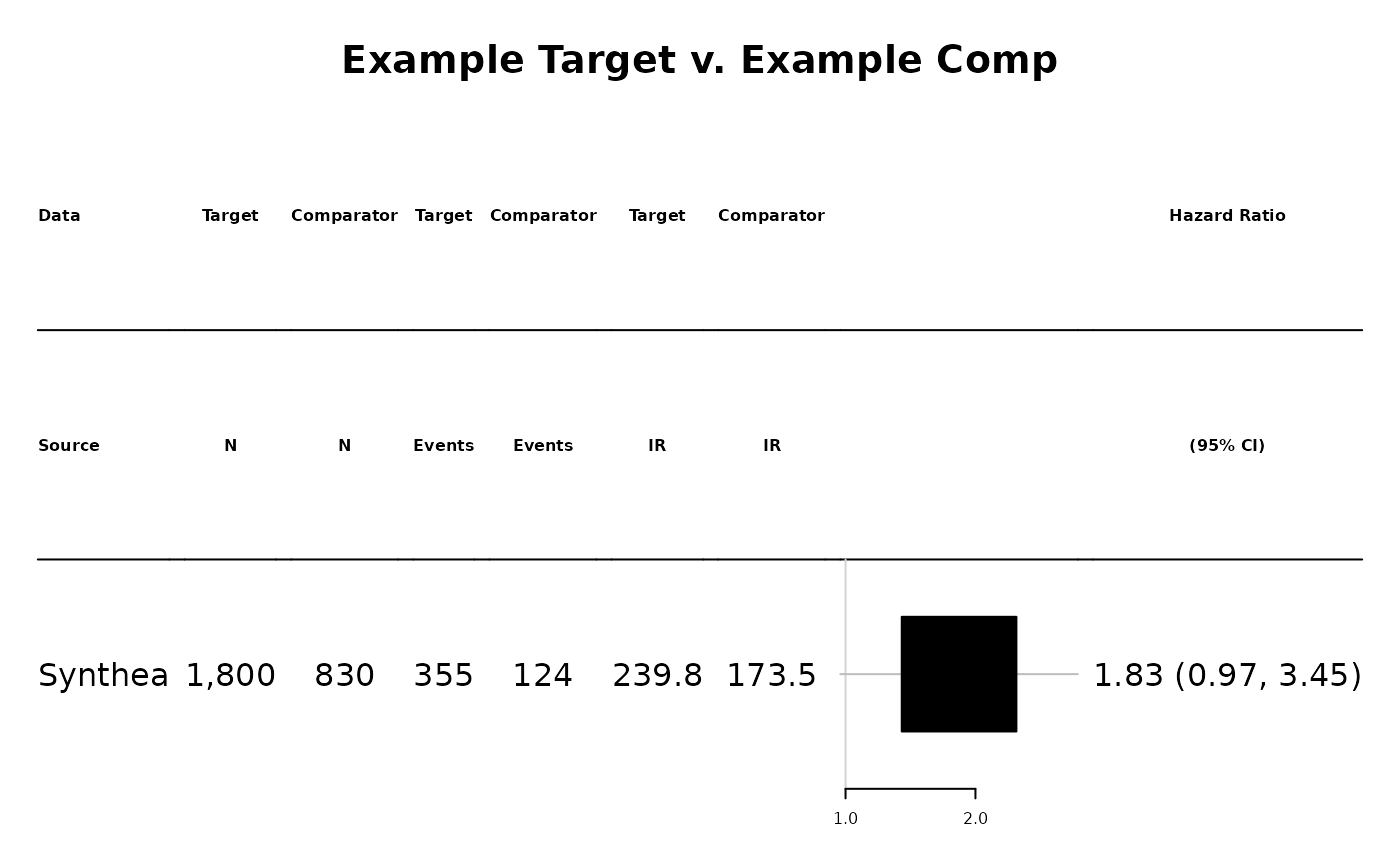Using Observational Health Data Sciences and Informatics (OHDSI) Report Generator
Jenna Reps, Anthony Sena
2026-02-06
Source:vignettes/ExampleCode.Rmd
ExampleCode.RmdUsing the OhdsiReportGenerator Package
This package enables users to extract characterization, estimation
and prediction results from the OHDSI results (see result
model) generated via R packages such as
PatientLevelPrediction, Characterization,
CohortMethod and SelfControlledCaseSeries.
Example Result Database
This package provides users with an example OHDSI result database.
This can be accessed via getExampleConnectionDetails() and
contains the results of running a demo analysis using the Eunomia R
package sample OMOP CDM data.
library(OhdsiReportGenerator)
# create a connection details object to
connectionDetails <- getExampleConnectionDetails()
# create a connection handler to the results
library(ResultModelManager)## Loading required package: R6## Loading required package: DatabaseConnector
ConnectionHandler <- ResultModelManager::ConnectionHandler$new(connectionDetails)## Connecting using SQLite driverExtracting Cohorts
The cohort details can be extracted using the function
getCohortDefinitions that requires the user to input the
connection handler, schema, table prefix used by CohortGenerator
(default is ‘cg_’) and target cohort ids to restrict to.
The example results are in a sqlite database and use the schema ‘main’.
cohorts <- getCohortDefinitions(
connectionHandler = ConnectionHandler,
schema = 'main',
cgTablePrefix = 'cg_',
targetIds = NULL
)
knitr::kable(
x = cohorts %>%
dplyr::select(-"json", -"sqlCommand"),
caption = 'The data.frame extracted containing the cohort details minus the json and sqlCommand columns.'
)| cohortDefinitionId | cohortName | description | subsetParent | isSubset | subsetDefinitionId | subsetDefinitionJson |
|---|---|---|---|---|---|---|
| 1 | Celecoxib | NA | 1 | NA | NA | NA |
| 2 | Diclofenac | NA | 2 | NA | NA | NA |
| 3 | GI bleed | NA | 3 | NA | NA | NA |
| 1001 | Celecoxib - age 18 to 64 Age 18 to 64 | NA | 1 | NA | 1 | { |
“name”: “age 18 to 64”, “definitionId”: 1, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 2001|Diclofenac - age 18 to 64 Age 18 to 64 |NA | 2| NA| 1|{ “name”: “age 18 to 64”, “definitionId”: 1, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 1002|Celecoxib - first event with 365 prior obs first event with 365 prior obs |NA | 1| NA| 2|{ “name”: “first event with 365 prior obs”, “definitionId”: 2, “subsetOperators”: [ { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 2002|Diclofenac - first event with 365 prior obs first event with 365 prior obs |NA | 2| NA| 2|{ “name”: “first event with 365 prior obs”, “definitionId”: 2, “subsetOperators”: [ { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 1003|Celecoxib - age 18 to 64 and first event with 365 prior obs Age 18 to 64, first event with 365 prior obs |NA | 1| NA| 3|{ “name”: “age 18 to 64 and first event with 365 prior obs”, “definitionId”: 3, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 }, { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 2003|Diclofenac - age 18 to 64 and first event with 365 prior obs Age 18 to 64, first event with 365 prior obs |NA | 2| NA| 3|{ “name”: “age 18 to 64 and first event with 365 prior obs”, “definitionId”: 3, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 }, { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } |
You can process the cohorts definitions to extract the parent cohorts and the children for each parent cohort.
if(nrow(cohorts) > 0){
processedCohorts <- processCohorts(cohorts)
knitr::kable(
x = data.frame(
parentId = processedCohorts$parents,
parentName= names(processedCohorts$parents)
),
caption = 'The parent cohorts.'
)
knitr::kable(
x = processedCohorts$cohortList[[1]] %>%
dplyr::select(-"json", -"sqlCommand"),
caption = 'The children/subset cohorts for the first parent cohort.'
)
}| cohortDefinitionId | cohortName | description | subsetParent | isSubset | subsetDefinitionId | subsetDefinitionJson |
|---|---|---|---|---|---|---|
| 1 | Celecoxib | NA | 1 | NA | NA | NA |
| 1001 | Celecoxib - age 18 to 64 Age 18 to 64 | NA | 1 | NA | 1 | { |
“name”: “age 18 to 64”, “definitionId”: 1, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 1002|Celecoxib - first event with 365 prior obs first event with 365 prior obs |NA | 1| NA| 2|{ “name”: “first event with 365 prior obs”, “definitionId”: 2, “subsetOperators”: [ { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 1003|Celecoxib - age 18 to 64 and first event with 365 prior obs Age 18 to 64, first event with 365 prior obs |NA | 1| NA| 3|{ “name”: “age 18 to 64 and first event with 365 prior obs”, “definitionId”: 3, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 }, { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } |
For example, if you created a cohort corresponding to all patients
exposed to drug A with an id of 1 and then created ‘children’ subset
cohorts corresponding to patients exposed to drug A for indication X
with an id 1001 and patients exposed to drug A aged 60+ with a cohort id
1002, then getCohortDefinition() would extract all three of
these cohorts and processCohorts() would identify cohort 1
as the parent and cohorts 1,1001,1002 as the children/subset of cohort
1.
You can also extract the subset logic for a subset id:
subsets <- getCohortSubsetDefinitions(
connectionHandler = ConnectionHandler,
schema = 'main',
cgTablePrefix = 'cg_'
)
knitr::kable(
x = subsets,
caption = 'The subset cohort logic used in the analysis.'
)| subsetDefinitionId | json |
|---|---|
| 1 | { |
“name”: “age 18 to 64”, “definitionId”: 1, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 2|{ “name”: “first event with 365 prior obs”, “definitionId”: 2, “subsetOperators”: [ { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } | | 3|{ “name”: “age 18 to 64 and first event with 365 prior obs”, “definitionId”: 3, “subsetOperators”: [ { “name”: “Age 18 to 64”, “subsetType”: “DemographicSubsetOperator”, “ageMin”: 18, “ageMax”: 64 }, { “name”: “first event with 365 prior obs”, “subsetType”: “LimitSubsetOperator”, “priorTime”: 365, “followUpTime”: 0, “limitTo”: “firstEver” } ], “packageVersion”: “0.11.2”, “identifierExpression”: “targetId * 1000 + definitionId”, “operatorNameConcatString”: “,”, “subsetCohortNameTemplate”: “@baseCohortName - @subsetDefinitionName @operatorNames” } |
Extracting Characterization Results
The characterization analysis contains time to event results, dechallenge-rechallenge, risk factor and cohort incidence results.
The time-to-event results tell you how often a outcome occurs relative to some target cohort index date on a 1-day, 30-day and 365-day scale.
tte <- getTimeToEvent(
connectionHandler = ConnectionHandler,
schema = 'main'
)
knitr::kable(
x = tte %>% dplyr::filter(.data$timeScale == 'per 365-day'),
caption = 'Example time-to-event results for the 365-day scale.'
)| databaseName | databaseId | targetName | targetId | outcomeName | outcomeId | outcomeType | targetOutcomeType | timeToEvent | numEvents | timeScale |
|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | first | After last target end | 365 | 327 | per 365-day |
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | first | Before first target start | 365 | 28 | per 365-day |
| Synthea | 388020256 | Diclofenac | 2 | GI bleed | 3 | first | After last target end | 365 | 114 | per 365-day |
| Synthea | 388020256 | Diclofenac | 2 | GI bleed | 3 | first | Before first target start | 365 | 10 | per 365-day |
| Synthea | 388020256 | Celecoxib - age 18 to 64 Age 18 to 64 | 1001 | GI bleed | 3 | first | After last target end | 365 | 327 | per 365-day |
| Synthea | 388020256 | Celecoxib - age 18 to 64 Age 18 to 64 | 1001 | GI bleed | 3 | first | Before first target start | 365 | 28 | per 365-day |
| Synthea | 388020256 | Diclofenac - age 18 to 64 Age 18 to 64 | 2001 | GI bleed | 3 | first | After last target end | 365 | 114 | per 365-day |
| Synthea | 388020256 | Diclofenac - age 18 to 64 Age 18 to 64 | 2001 | GI bleed | 3 | first | Before first target start | 365 | 10 | per 365-day |
The dechallenge-rechallenge analysis shows you how often an outcome occurs during some target exposure and then how often the target exposure is stopped shortly after the outcome and whether the outcome re-occurs when the target exposure restarts:
drc <- getDechallengeRechallenge(
connectionHandler = ConnectionHandler,
schema = 'main'
)
knitr::kable(
x = drc,
caption = 'Example dechallenge-rechallenge results.'
)| databaseName | databaseId | targetName | targetId | outcomeName | outcomeId | dechallengeStopInterval | dechallengeEvaluationWindow | numExposureEras | numPersonsExposed | numCases | dechallengeAttempt | dechallengeFail | dechallengeSuccess | rechallengeAttempt | rechallengeFail | rechallengeSuccess | pctDechallengeAttempt | pctDechallengeFail | pctDechallengeSuccess | pctRechallengeAttempt | pctRechallengeFail | pctRechallengeSuccess |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | 30 | 30 | 100 | 80 | 20 | 15 | 10 | 5 | 5 | 1 | 4 | 0.75 | 0.67 | 0.33 | 1 | 0.2 | 0.8 |
It is also possible to find the incidence rates (where we restrict to all ages, genders and start years):
ir <- getIncidenceRates(
connectionHandler = ConnectionHandler,
schema = 'main',
targetIds = 1
)
knitr::kable(
x = ir %>% dplyr::filter(
subgroupName == 'All' &
ageGroupName == 'Any' &
genderName == 'Any' &
startYear == 'Any'
),
caption = 'Example incidence rate results.'
)| databaseName | databaseId | targetName | targetId | outcomeName | outcomeId | tar | cleanWindow | subgroupName | ageGroupName | genderName | startYear | tarStartWith | tarStartOffset | tarEndWith | tarEndOffset | personsAtRiskPe | personsAtRisk | personDaysPe | personDays | personOutcomesPe | personOutcomes | outcomesPe | outcomes | incidenceProportionP100p | incidenceRateP100py |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | ( start + 0 ) - ( end + 0 ) | 9999 | All | Any | Any | Any | start | 0 | end | 0 | 1800 | 1800 | 1800 | 1800 | 0 | 0 | 0 | 0 | 0 | 0 |
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | ( start + 0 ) - ( start + 365 ) | 9999 | All | Any | Any | Any | start | 0 | start | 365 | 1800 | 1800 | 648214 | 540789 | 0 | 0 | 0 | 0 | 0 | 0 |
Finally, it is possible to get risk factors (associations between
features and the occurrence of the outcome during a time-at-risk) using
getBinaryRiskFactors to identify the binary features:
rf <- getBinaryRiskFactors(
connectionHandler = ConnectionHandler,
schema = 'main',
targetId = 1,
outcomeId = 3
)
knitr::kable(
x = rf,
caption = 'Example risk factors for binary features results.'
)| databaseName | databaseId | targetName | targetCohortId | outcomeName | outcomeCohortId | minPriorObservation | outcomeWashoutDays | riskWindowStart | riskWindowEnd | startAnchor | endAnchor | covariateName | covariateId | caseCount | caseAverage | nonCaseCount | nonCaseAverage | SMD | absSMD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | 365 | 365 | 1 | 365 | cohort start | cohort end | age group: 30 - 34 | 6003 | 35 | 0.0985916 | 171 | 0.1183391 | -0.0635357 | 0.0635357 |
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | 365 | 365 | 1 | 365 | cohort start | cohort end | age group: 35 - 39 | 7003 | 177 | 0.4985915 | 685 | 0.4740484 | 0.0789649 | 0.0789649 |
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | 365 | 365 | 1 | 365 | cohort start | cohort end | age group: 40 - 44 | 8003 | 129 | 0.3633803 | 531 | 0.3674740 | -0.0131713 | 0.0131713 |
| Synthea | 388020256 | Celecoxib | 1 | GI bleed | 3 | 365 | 365 | 1 | 365 | cohort start | cohort end | age group: 45 - 49 | 9003 | 14 | 0.0394366 | 58 | 0.0401384 | -0.0022579 | 0.0022579 |
and getContinuousRiskFactors for the continuous
features:
rf <- getContinuousRiskFactors(
connectionHandler = ConnectionHandler,
schema = 'main',
targetId = 1,
outcomeId = 3
)
knitr::kable(
x = rf,
caption = 'Example risk factors for continuous features results.'
)| databaseName | databaseId | targetName | targetCohortId | minPriorObservation | covariateName | covariateId | targetCountValue | targetMinValue | targetMaxValue | targetAverageValue | targetStandardDeviation | targetMedianValue | targetP10Value | targetP25Value | targetP75Value | targetP90Value | outcomeName | outcomeCohortId | outcomeWashoutDays | riskWindowStart | riskWindowEnd | startAnchor | endAnchor | caseCountValue | caseMinValue | caseMaxValue | caseAverageValue | caseStandardDeviation | caseMedianValue | caseP10Value | caseP25Value | caseP75Value | caseP90Value | SMD | absSMD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | Celecoxib | 1 | 365 | age in years | 1002 | 1800 | 31 | 47 | 38.645 | 3.321244 | 39 | 34 | 36 | 41 | 43 | GI bleed | 3 | 365 | 1 | 365 | cohort start | cohort end | 355 | 32 | 46 | 38.77465 | 3.274612 | 39 | 35 | 36 | 41 | 43 | 0.0393116 | 0.0393116 |
| Synthea | 388020256 | Celecoxib | 1 | 365 | observation time (days) after index | 1009 | 1800 | 10 | 28328 | 7588.082 | 5352.386801 | 6676 | 1336 | 3442 | 10649 | 14808 | GI bleed | 3 | 365 | 1 | 365 | cohort start | cohort end | 355 | 10 | 27390 | 7524.37465 | 5611.529121 | 6708 | 996 | 3236 | 10415 | 14884 | -0.0116180 | 0.0116180 |
| Synthea | 388020256 | Celecoxib | 1 | 365 | observation time (days) prior to index | 1008 | 1800 | 11369 | 17044 | 14114.684 | 1195.708067 | 14100 | 12544 | 13160 | 15040 | 15744 | GI bleed | 3 | 365 | 1 | 365 | cohort start | cohort end | 355 | 11550 | 16805 | 14159.86761 | 1176.652643 | 14155 | 12644 | 13250 | 15075 | 15840 | 0.0380906 | 0.0380906 |
| Synthea | 388020256 | Celecoxib | 1 | 365 | time (days) between cohort start and end | 1010 | 1800 | 0 | 0 | 0.000 | 0.000000 | 0 | 0 | 0 | 0 | 0 | GI bleed | 3 | 365 | 1 | 365 | cohort start | cohort end | 355 | 0 | 0 | 0.00000 | 0.000000 | 0 | 0 | 0 | 0 | 0 | NaN | NaN |
| Synthea | 388020256 | Celecoxib | 1 | 365 | visit_occurrence concept count during day -365 through 0 concept_count relative to index: Inpatient Visit | 9201923 | -5 | 0 | 0 | 0.000 | 0.000000 | 0 | 0 | 0 | 0 | 0 | GI bleed | 3 | 365 | 1 | 365 | cohort start | cohort end | 0 | 0 | 0 | 0.00000 | 0.000000 | 0 | 0 | 0 | 0 | 0 | NaN | NaN |
Extracting Prediction Results
The model designs used to develop prediction models in an analysis
can be extracted using getPredictionModelDesigns:
modelDesigns <- getPredictionModelDesigns(
connectionHandler = ConnectionHandler,
schema = 'main',
targetIds = 1002,
outcomeIds = 3
)
knitr::kable(
x = modelDesigns %>%
dplyr::select(
"modelDesignId",
"modelType",
"developmentTargetId",
"developmentTargetName",
"developmentOutcomeId",
"developmentOutcomeName",
"timeAtRisk"
),
caption = 'Example model designs for the prediction results.'
)| modelDesignId | modelType | developmentTargetId | developmentTargetName | developmentOutcomeId | developmentOutcomeName | timeAtRisk |
|---|---|---|---|---|---|---|
| 1 | logistic | 1002 | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 3 | GI bleed | (cohort start + 1) - (cohort start + 365) |
Once you know the modelDesignId of interest, you can then extract the model performance:
perform <- getFullPredictionPerformances(
connectionHandler = ConnectionHandler,
schema = 'main',
modelDesignId = 1
)
knitr::kable(
x = perform,
caption = 'Example performance for the prediction results.'
)| performanceId | modelDesignId | timeStamp | modelType | covariateName | developmentDatabaseId | validationDatabaseId | developmentTargetId | developmentTargetName | validationTargetId | validationTargetName | developmentOutcomeId | developmentOutcomeName | validationOutcomeId | validationOutcomeName | developmentDatabase | validationDatabase | validationTarId | developmentTarId | validationTimeAtRisk | developmentTimeAtRisk | evaluation | populationSize | outcomeCount | AUROC | 95% lower AUROC | 95% upper AUROC | AUPRC | brier score | brier score scaled | Eavg | E90 | Emax | calibrationInLarge mean prediction | calibrationInLarge observed risk | calibrationInLarge intercept | weak calibration intercept | weak calibration gradient | Hosmer-Lemeshow calibration intercept | Hosmer-Lemeshow calibration gradient | Average Precision |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 2025-02-13 | logistic | getDbDefaultCovariateData (29 types) inc age and sex | 1 | 1 | 1002 | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 1002 | - | 3 | GI bleed | 3 | - | Synthea | - | 1 | 1 | (cohort start + 1) - (cohort start + 365) | (cohort start + 1) - (cohort start + 365) | Test | 449 | 88 | 0.6885703 | 0.6264985 | 0.7506421 | 0.3085693 | 0.1469204 | 0.0720927 | 0.0318938 | 0.0650422 | 0.1390867 | 0.1972381 | 0.1959911 | 0.0976443 | 0.0976443 | 1.081961 | 0.0150102 | 0.9957815 | 0.3141461 |
| 1 | 1 | 2025-02-13 | logistic | getDbDefaultCovariateData (29 types) inc age and sex | 1 | 1 | 1002 | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 1002 | - | 3 | GI bleed | 3 | - | Synthea | - | 1 | 1 | (cohort start + 1) - (cohort start + 365) | (cohort start + 1) - (cohort start + 365) | Train | 1351 | 267 | 0.7091712 | 0.6750978 | 0.7432446 | 0.3665755 | 0.1451619 | 0.0845800 | 0.0211919 | 0.0365588 | 0.0525081 | 0.1976328 | 0.1976314 | 0.2335147 | 0.2335147 | 1.181563 | -0.0358802 | 1.1620115 | 0.3702857 |
| 1 | 1 | 2025-02-13 | logistic | getDbDefaultCovariateData (29 types) inc age and sex | 1 | 1 | 1002 | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 1002 | - | 3 | GI bleed | 3 | - | Synthea | - | 1 | 1 | (cohort start + 1) - (cohort start + 365) | (cohort start + 1) - (cohort start + 365) | CV | 1351 | 267 | 0.6818760 | 0.6457077 | 0.7180443 | 0.3151896 | 0.1490119 | 0.0586252 | 0.0265908 | 0.0447049 | 0.2297922 | 0.1971663 | 0.1976314 | 0.0905528 | 0.0905528 | 1.066780 | -0.0116256 | 1.0545190 | 0.3196781 |
As well as the top predictors, in this case we will return the top 5:
top5 <- getPredictionTopPredictors(
connectionHandler = ConnectionHandler,
schema = 'main',
targetIds = 1002,
outcomeIds = 3,
numberPredictors = 5
)
knitr::kable(
x = top5 ,
caption = 'Example top five predictors.'
)| databaseName | tarStartDay | tarStartAnchor | tarEndDay | tarEndAnchor | performanceId | covariateId | covariateName | conceptId | covariateValue | covariateCount | covariateMean | covariateStDev | withNoOutcomeCovariateCount | withNoOutcomeCovariateMean | withNoOutcomeCovariateStDev | withOutcomeCovariateCount | withOutcomeCovariateMean | withOutcomeCovariateStDev | standardizedMeanDiff | rn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 1 | cohort start | 365 | cohort start | 1 | 1901 | Charlson index - Romano adaptation | 0 | 1.4448410 | 935 | 0.6144444 | 0.6533777 | 660 | 0.5307958 | 0.6301972 | 275 | 0.9549296 | 0.6352971 | 0.6702998 | 1 |
| Synthea | 1 | cohort start | 365 | cohort start | 1 | 4266809210 | condition_era group (ConditionGroupEraLongTerm) during day -365 through 0 days relative to index: Diverticular disease | 4266809 | 0.5045103 | 338 | 0.1877778 | 0.3905346 | 210 | 0.1453287 | 0.3524320 | 128 | 0.3605634 | 0.4801640 | 0.5110418 | 2 |
| Synthea | 1 | cohort start | 365 | cohort start | 1 | 4285898212 | condition_era group (ConditionGroupEraShortTerm) during day -30 through 0 days relative to index: Polyp of colon | 4285898 | 0.4321971 | 28 | 0.0155556 | 0.1237481 | 13 | 0.0089965 | 0.0944225 | 15 | 0.0422535 | 0.2011670 | 0.2116439 | 3 |
| Synthea | 1 | cohort start | 365 | cohort start | 1 | 2007 | index month: 2 | 0 | -0.2439688 | 124 | 0.0688889 | 0.2532651 | 104 | 0.0719723 | 0.2584421 | 20 | 0.0563380 | 0.2305733 | -0.0638383 | 4 |
| Synthea | 1 | cohort start | 365 | cohort start | 1 | 6007 | index month: 6 | 0 | -0.1786601 | 144 | 0.0800000 | 0.2712932 | 122 | 0.0844291 | 0.2780302 | 22 | 0.0619718 | 0.2411044 | -0.0862999 | 5 |
You can also select certain results such as the attrition:
attrition <- getPredictionPerformanceTable(
connectionHandler = ConnectionHandler,
schema = 'main',
table = 'attrition',
performanceId = 1
)
knitr::kable(
x = attrition ,
caption = 'Example prediction attrition for performance 1.'
)| modelDesignId | developmentDatabaseId | developmentDatabaseName | validationDatabaseId | validationDatabaseName | targetId | outcomeId | tarId | performanceId | outcomeId | description | targetCount | uniquePeople | outcomes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Synthea | 1 | Synthea | 1 | 2 | 1 | 1 | 3 | Original cohorts | 1800 | 1800 | 355 |
| 1 | 1 | Synthea | 1 | Synthea | 1 | 2 | 1 | 1 | 3 | Initial plpData cohort or population | 1800 | 1800 | 355 |
| 1 | 1 | Synthea | 1 | Synthea | 1 | 2 | 1 | 1 | 3 | No prior outcome | 1800 | 1800 | 355 |
| 1 | 1 | Synthea | 1 | Synthea | 1 | 2 | 1 | 1 | 3 | Removing non-outcome subjects with insufficient time at risk (if any) | 1800 | 1800 | 355 |
Extracting Estimation Results
It is possible to extract cohort method and self controlled case series results and make nice plots.
To extract all the cohort method estimations for target id 1002 and outcome 3:
cmEst <- getCMEstimation(
connectionHandler = ConnectionHandler,
schema = 'main',
targetIds = 1002,
comparatorIds = 2002,
outcomeIds = 3
)
knitr::kable(
x = cmEst ,
caption = 'Example cohort method estimation results.'
)| databaseName | databaseId | analysisId | description | targetName | targetId | comparatorName | comparatorId | outcomeName | outcomeId | calibratedRr | calibratedCi95Lb | calibratedCi95Ub | calibratedP | calibratedOneSidedP | calibratedLogRr | calibratedSeLogRr | targetSubjects | comparatorSubjects | targetDays | comparatorDays | targetOutcomes | comparatorOutcomes | unblind | unblindForEvidenceSynthesis | targetEstimator |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthea | 388020256 | 1 | No matching, simple outcome model | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 1002 | Diclofenac - first event with 365 prior obs first event with 365 prior obs | 2002 | GI bleed | 3 | 1.832771 | 0.9724055 | 3.454372 | 0.1078385 | 0.0639169 | 0.6058291 | 0.3233791 | 1800 | 830 | 540789 | 261005 | 355 | 124 | 1 | 1 | ate |
| Synthea | 388020256 | 2 | Matching on ps and covariates, simple outcomeModel | Celecoxib - first event with 365 prior obs first event with 365 prior obs | 1002 | Diclofenac - first event with 365 prior obs first event with 365 prior obs | 2002 | GI bleed | 3 | NA | NA | NA | NA | NA | NA | NA | 829 | 829 | 263258 | 260639 | 117 | 124 | 0 | 0 | att |
You can also extract any meta-analysis estimate for cohort method:
cmMe <- getCmMetaEstimation(
connectionHandler = ConnectionHandler,
schema = 'main'
)
knitr::kable(
x = cmMe ,
caption = 'Example cohort method meta analysis estimation results.'
)| databaseName | analysisId | description | targetName | targetId | comparatorName | comparatorId | outcomeName | outcomeId | calibratedRr | calibratedCi95Lb | calibratedCi95Ub | calibratedP | calibratedOneSidedP | calibratedLogRr | calibratedSeLogRr | targetSubjects | comparatorSubjects | targetDays | comparatorDays | targetOutcomes | comparatorOutcomes | unblind | nDatabases |
|---|
and you can create a nice plot to show the results:
plotCmEstimates(
cmData = cmEst,
cmMeta = NULL,
targetName = 'Example Target',
comparatorName = 'Example Comp',
selectedAnalysisId = 1
)