
Create a plot from the output of summariseObservationPeriod()
Source:R/plotObservationPeriod.R
plotObservationPeriod.RdCreate a plot from the output of summariseObservationPeriod()
Usage
plotObservationPeriod(
result,
variableName = "Number subjects",
plotType = "barplot",
facet = NULL,
colour = NULL,
style = NULL,
type = NULL
)Arguments
- result
A summarised_result object (output of
summariseObservationPeriod()).- variableName
The variable to plot it can be: "Number subjects", "Records per person", "Duration in days" or "Days to next observation period".
- plotType
The plot type, it can be: "barplot", "boxplot" or "densityplot".
- facet
Columns to face by. Formula format can be provided. See possible columns to face by with:
visOmopResults::tidyColumns().- colour
Columns to colour by. See possible columns to colour by with:
visOmopResults::tidyColumns().- style
Visual theme to apply. Character, or
NULL. If a character, this may be either the name of a built-in style (seevisOmopResults::plotStyle()), or a path to a .yml file that defines a custom style. IfNULL, the function will use the explicit default style, unless a global style option is set (seevisOmopResults::setGlobalPlotOptions()) or a _brand.yml file is present (in that order).- type
Character string indicating the output plot format. See
visOmopResults::plotType()for the list of supported plot types. Iftype = NULL, the function will use the global setting defined viavisOmopResults::setGlobalPlotOptions()(if available); otherwise, a standardggplot2plot is produced by default.
Examples
# \donttest{
library(OmopSketch)
library(dplyr, warn.conflicts = FALSE)
library(omock)
cdm <- mockCdmFromDataset(datasetName = "GiBleed", source = "duckdb")
#> ℹ Loading bundled GiBleed tables from package data.
#> ℹ Adding drug_strength table.
#> ℹ Creating local <cdm_reference> object.
#> ℹ Inserting <cdm_reference> into duckdb.
result <- summariseObservationPeriod(cdm = cdm)
#> Warning: ! There are 2649 individuals not included in the person table.
tableObservationPeriod(result = result)
Summary of observation_period table
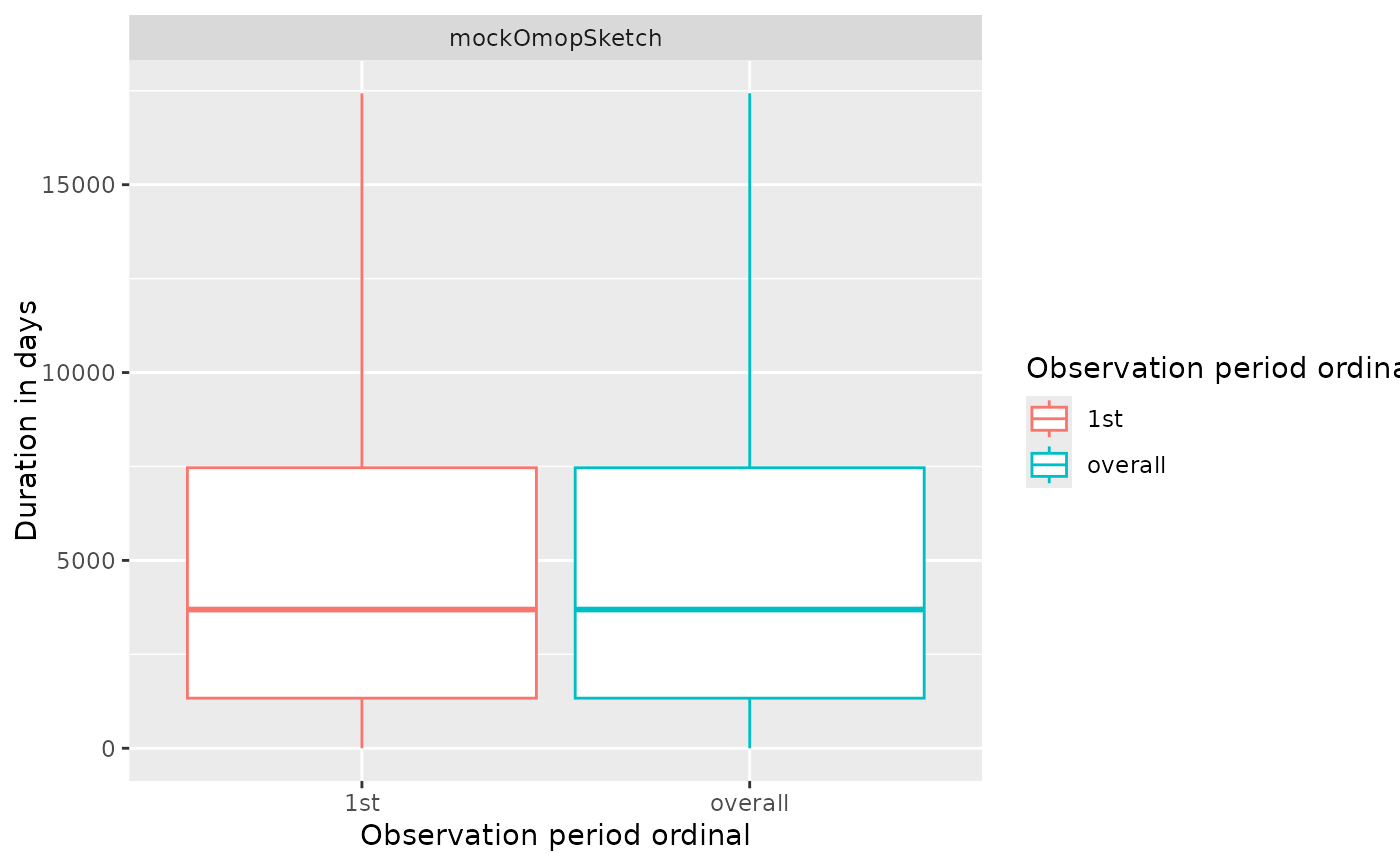
plotObservationPeriod(
result = result,
variableName = "Duration in days",
plotType = "boxplot"
)
 cdmDisconnect(cdm = cdm)
# }
cdmDisconnect(cdm = cdm)
# }