 OHDSI GIS WG Geocoding Workflow
OHDSI GIS WG Geocoding Workflow
 OHDSI GIS WG Geocoding Workflow
OHDSI GIS WG Geocoding Workflow
Overview
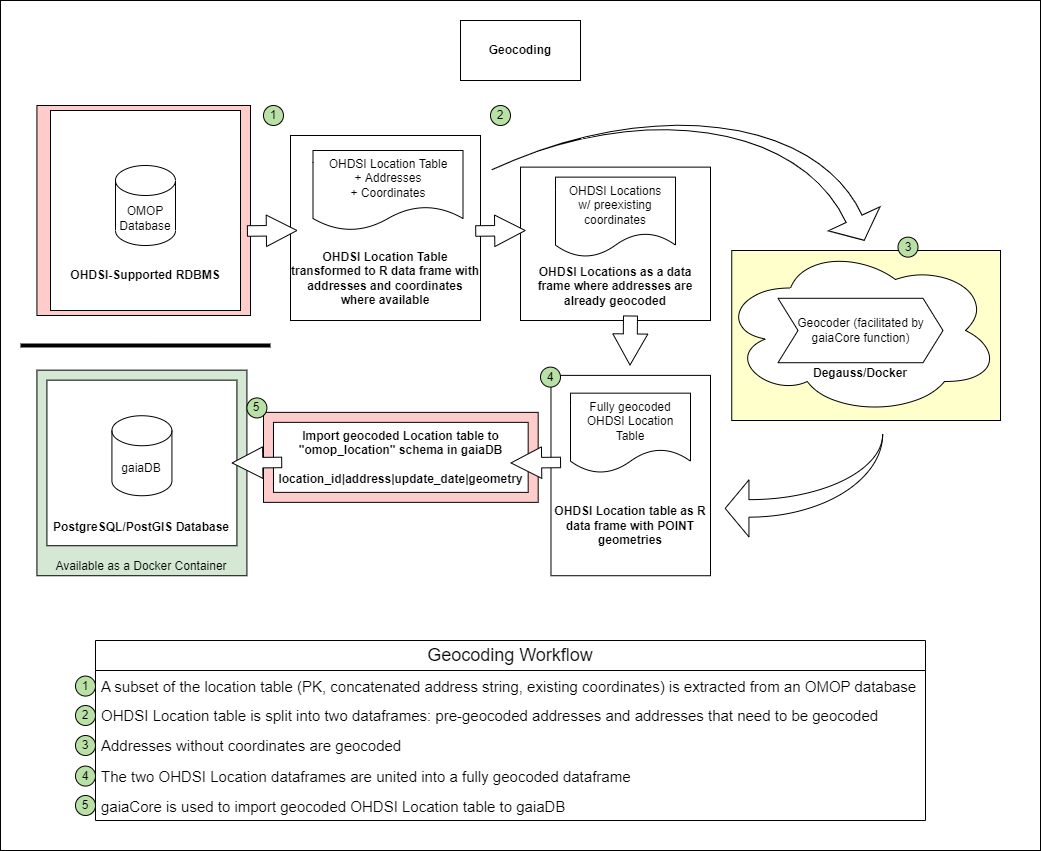
This workflow demonstrates how to use the OHDSI Gaia suite of GIS tools to geocode the entirety of an OMOP Location table, fully enabling any further geospatial analyses supported by the Gaia GIS tools.
Geocoding Workflow (Cohort Definitions) - Click here to download
Prerequisites
This workflow requires the DatabaseConnector
package.
install.packages("DatabaseConnector")
library(DatabaseConnector)Configuring Connection to the Servers
We need to configure connections to two servers: the server hosting the OMOP database and the server hosting the gaiaDB Postgres database.
Connect to OMOP server:
connectionDetails <- createConnectionDetails(
dbms = keyring::key_get('cdm-dbms'),
user = keyring::key_get('cdm-username'),
password = keyring::key_get('cdm-password'),
server = keyring::key_get('cdm-server')
)
cdmDatabaseSchema <- "TMC_RED.dbo"Connect to gaiaDB server:
If you don’t already have a gaiaDB server set up, see the installation instructions before proceeding.
library(DatabaseConnector)
gaiaConnectionDetails <- DatabaseConnector::createConnectionDetails(
dbms = "postgresql",
server = "localhost/gaiaDB",
port = 5432,
user="postgres",
password = "mySecretPassword") Geocoding
Step 1. Ingest the Location table from OMOP Database
Use the gaiaCore function getLocationAddresses() to
extract addresses from OMOP and return a transformed location table:
transformedLocation <- getLocationAddresses(connectionDetails = connectionDetails,
cdmDatabaseSchema = cdmDatabaseSchema)Step 2. Split the transformedLocation dataframe into two tables based on whether it already contains latitude and longitude information:
splitResult <- splitAddresses(addressTable = transformedLocation)
alreadyGeocodedLocation <- splitResult$geocoded
notGeocodedLocation <- splitResult$ungeocodedStep 3. Geocode Addresses without Coordinates
geocodedLocation <- geocodeAddresses(addressTable = notGeocodedLocation)Step 4. Stack pre-populated and newly geocoded location tables
fullyGeocodedLocation <- geocodedLocation
if (!is.null(alreadyGeocodedLocation)) {
names(alreadyGeocodedLocation) <- tolower(names(alreadyGeocodedLocation))
alreadyGeocodedLocation <- dplyr::mutate(alreadyGeocodedLocation, lat = latitude, lon = longitude)
alreadyGeocodedLocation <- dplyr::select(alreadyGeocodedLocation, names(fullyGeocodedLocation))
alreadyGeocodedLocation <- sf::st_as_sf(boundGeocodedTable, coords = c("lon", "lat"), crs = 4326)
fullyGeocodedLocation <- rbind(fullyGeocodedLocation, alreadyGeocodedLocation)
}Step 5. Import Geocoded Location table to gaiaDB Database
importLocationTable(gaiaConnectionDetails = gaiaConnectionDetails,
location = fullyGeocodedLocation) OHDSI GIS
OHDSI GIS