CDM Table: STEM
The STEM table is a staging area where source codes like ICD9 codes will first be mapped to concept_ids. The STEM table itself is an amalgamation of the OMOP event tables to facilitate record movement. This means that all fields present across the OMOP event tables are present in the STEM table. After a record is mapped and staged, the domain of the concept_id dictates which OMOP table (Condition_occurrence, Drug_exposure, Procedure_occurrence, Measurement, Observation, Device_exposure) the record will move to. Please see the STEM -> CDM mapping files for a description of which STEM fields move to which STEM tables.
Notes
- VISIT_DETAIL must be built before STEM (refer to VISIT_DETAIL file)
- The MED_PROCEDURE table can be joined to MEDICAL_CLAIMS which is joined to VISIT_DETAIL.
- Referential integrity is maintained with VISIT_DETAIL.
- For every record in STEM there should be 1 row record in VISIT_DETAIL (n:1 join).
- For every record in VISIT_DETAIL there may be 0 to n rows in STEM.
Mapping from LAB_RESULTS
- Take the records from LAB_RESULTS and join them to MEDICAL_CLAIMS on - patid = patid - pat_planid = pat_planid - labclmid = clmid
- This will assign each lab result at least one VISIT_DETAIL VISIT_DETAIL_ID based on the lookup created during VISIT_DETAIL creation. If more than one is assigned, choose one.
- Some records in LAB_RESULTS do not have a labclmid. If this is the case PROCEDURE_OCCURRENCE VISIT_DETAIL_ID and VISIT_OCCURRENCE_ID should be left blank.
- This will assign each lab result at least one VISIT_DETAIL VISIT_DETAIL_ID based on the lookup created during VISIT_DETAIL creation. If more than one is assigned, choose one.
- RSLT_NBR will go to VALUE_AS_NUMBER and RSLT_TXT and RSLT_NBR will be concatenated in the VALUE_SOURCE_VALUE field.
- RSLT_TXT should then be mapped to a concept by looking up the result value in the LOINC vocabulary
- The RSLT_TXT field tends to have operators (<, >, =, ≤, >=) included in the first 2 characters of the string. Use the following logic to assign the OPERATOR_CONCEPT_ID in such cases:
case substring(rslt_txt, 0, 2) when '<' then operator_concept_id = 4172704 when '>' then operator_concept_id = 4171756 when '=' then operator_concept_id = 4172703 when '>=' then operator_concept_id = 4171755 when '<=' then operator_concept_id = 4171754 end
- START_DATE is assigned to VISIT_DETAIL_START_DATE
- The VISIT_DETAIL.VISIT_OCCURRENCE_ID and VISIT_DETAIL.VISIT_DETAIL_ID are FK in STEM table.
Mapping COVID-19 test result text values
Certain COVID-19 tests have specific text results that need to be mapped manually to Standard Concepts in the VALUE_AS_CONCEPT field. If any of the below values show up in the RSLT_TXT field, map them to concepts using the below table.
| RSTL_TXT Value | VALUE_AS_CONCEPT_ID |
|---|---|
| LDTNOT | 9190 |
| NEG | 9190 |
| Not-Detected | 9190 |
| NOTDET | 9190 |
| Not Detected^Not D | 9190 |
| Negative for COVID | 9190 |
| LDTDET | 4126681 |
| POS | 4126681 |
| Positive for 2019- | 4126681 |
| Positive for COVID | 4126681 |
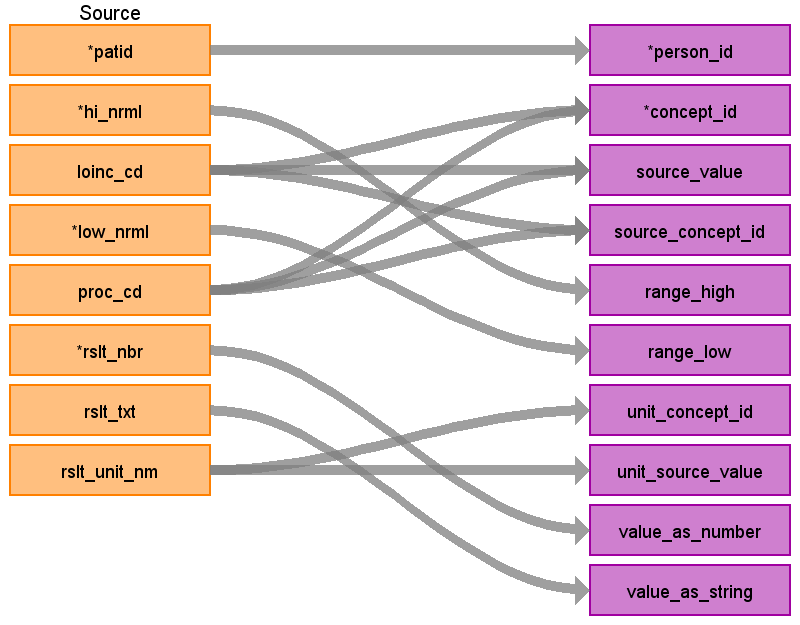
| Destination Field | Source Field | Applied Rule | Comment | ||
|---|---|---|---|---|---|
| id | Autogenerate | ||||
| domain_id | This should be the domain_id of the standard concept in the CONCEPT_ID field. If a code is mapped to CONCEPT_ID 0, put the domain_id as Observation. | ||||
| person_id | VISIT_DETAIL PERSON_ID LAB_RESULT PATID | Use patid to lookup Person_id | |||
| visit_detail_id | VISIT_DETAIL VISIT_DETAIL_ID | ||||
| visit_occurrence_id | VISIT_DETAIL VISIT_OCCURRENCE_ID | Use the linking to VISIT_DETAIL to look up VISIT_OCCURRENCE_ID | |||
| provider_id | VISIT_DETAIL PROVIDER_ID | ||||
| start_datetime | VISIT_DETAIL VISIT_DETAIL_START_DATETIME | ||||
| concept_id | LOINC_CD, PROC_CD | Use the SOURCE_TO_STANDARD query with the filter LOINC_CD WHERE SOURCE_VOCABULARY_ID IN (‘LOINC’) AND TARGET_STANDARD_CONCEPT =’S’ AND TARGET_INVALID_REASON IS NULL PROC_CD WHERE SOURCE_VOCABULARY_ID IN (‘HCPCS’,’CPT4’) AND TARGET_STANDARD_CONCEPT =’S’ AND TARGET_INVALID_REASON IS NULL AND TARGET_CONCEPT_CLASS_ID NOT IN (‘HCPCS Modifier’,’CPT4 Modifier’) | Start with LOINC_CD, if it does not have a mapping then try to map the PROC_CD. If the PROC_CD does not have a mapping then set concept_id to 0 | ||
| source_value | LOINC_CD, PROC_CD | ||||
| source_concept_id | LOINC_CD, PROC_CD | Use the SOURCE_TO_SOURCE query with the filter LOINC_CD WHERE SOURCE_VOCABULARY_ID IN (‘LOINC’) PROC_CD WHERE SOURCE_VOCABULARY_ID IN (‘HCPCS’,’CPT4’) | Put the SOURCE_CONCEPT_ID of the either the LOINC_CD or PROC_CD that was used to map the standard concept_id. | ||
| type_concept_id | Set to 32856 (Lab) | ||||
| operator_concept_id | See the above logic to assign this value based on the first to characters of the RSLT_TXT field. | ||||
| unit_concept_id | LAB_RESULTS RSLT_UNIT_NM | Use the SOURCE_TO_STANDARD query with the filter Where SOURCE_VOCABULARY_ID in (‘UCUM’, ‘JNJ_UNITS’) AND TARGET_STANDARD_CONCEPT = ‘S’ AND TARGET_INVALID_REASON IS NULL | If there is no mapping to a standard concept then set to 0 Set UNIT_CONCEPT_ID = NULL when the source unit value is NULL | ||
| unit_source_value | LAB_RESULTS RSLT_UNIT_NM | ||||
| start_date | VISIT_DETAIL VISIT_DETAIL_START_DATE | ||||
| end_date | |||||
| range_high | LAB_RESULTS HI_NRML | ||||
| range_low | LAB_RESULTS LOW_NRML | ||||
| value_as_number | LAB_RESULTS RSLT_NBR | ||||
| value_as_string | |||||
| value_as_concept_id | LAB_RESULTS RSLT_TXT | Use the SOURCE_TO_STANDARD query with the filter LOINC_CD WHERE SOURCE_VOCABULARY_ID IN (‘LOINC’) AND TARGET_STANDARD_CONCEPT =’S’ AND TARGET_INVALID_REASON IS NULL, mapping to SOURCE_CODE_DESCRIPTION instead of SOURCE_CODE | |||
| value_source_value | LAB_RESULTS RSLT_NBR, RSLT_TXT | Concatenate RSLT_NBR, RSLT_TXT with ‘;’ between. | |||
| end_datetime | |||||
| verbatim_end_date | |||||
| days_supply | |||||
| dose_unit_source_value | |||||
| lot_number | |||||
| MODIFIER_CONCEPT_ID | |||||
| modifier_source_value | |||||
| quantity | |||||
| refills | |||||
| route_concept_id | |||||
| route_source_value | |||||
| sig | |||||
| stop_reason | |||||
| unique_device_id | |||||
| anatomic_site_concept_id | |||||
| disease_status_concept_id | |||||
| specimen_source_id | |||||
| anatomic_site_source_value | |||||
| disease_status_source_value | |||||
| condition_status_concept_id | |||||
| condition_status_source_value |
Change Log
October 15, 2021
- Added JNJ_UNITS as a vocabulary to use when mapping UNIT_CONCEPT_ID
July 14, 2021
- Changed RSLT_TXT from going to VALUE_AS_STRING to VALUE_SOURCE_VALUE
- Added a mapping for VALUE_AS_CONCEPT_ID
- Added logic to map OPERATOR_CONCEPT_ID